Gene
KWMTBOMO02936 Validated by peptides from experiments
Annotation
uncharacterized_protein_LOC778510_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 3.547
Sequence
CDS
ATGGCACAAACAGCGGGTGTTAAACCTATGACGATAGTTGGTCGCGTAGCCAGTGAAAGGGAAAGATGTCTTGGTATGACTGATGCAGAAAGGGCTTGGCGTAAGCAATGGTTAAAAGATCAAGTTTTAGCCGCTCACGAGCCAGTTCACGTGGAAGAATATTGGAGAGAGCGTACTAATCCTATTCGTCGTTTTTACCGGAAACCCTTGGACGTACTCTTTGCGAAACTTACCCCAATGCTGGGAGAACAACGTGCTGCACATTATAGATATATATCTGGAAAACTTGGTTTAATCGCTGTGGCTATGCTATCAACACATTACTACTTTAAATATTTAGGAAACGACTGGACAAAGAAGGGTGGTTGGAAAGTACTAAAAACCAAACCAATGGTTCTTCCTGGACAACCTGGATTCCCATTCAAATCTGAGAAAACCGATTCTGATTACGCTGAAAGGAAATTCAAGAGCTCTGTTATTTAA
Protein
MAQTAGVKPMTIVGRVASERERCLGMTDAERAWRKQWLKDQVLAAHEPVHVEEYWRERTNPIRRFYRKPLDVLFAKLTPMLGEQRAAHYRYISGKLGLIAVAMLSTHYYFKYLGNDWTKKGGWKVLKTKPMVLPGQPGFPFKSEKTDSDYAERKFKSSVI
Summary
Uniprot
A0FDR2
S4NNG4
A0A212FF15
A0A0N1IPM2
I4DKI2
A0A194PNP8
+ More
A0A2H1V7Z2 A0A2W1BRZ6 A0A2A4K4Z6 A0A0L7KNJ0 A0A1L8E162 A0A023EG32 A0A182H4X5 W5JUX8 A0A182MHR1 A0A2M4CV22 A0A2M3Z495 A0A2M4C1L4 T1DJ84 A0A2M4AFC2 A0A182WHI5 A0A182RRI5 A0A1Q3FIX2 A0A182NUW8 A0A2J7R2Q2 A0A182P3C2 A0A1B0D6Z1 B0W0C0 Q16WX6 A0A182Y5L0 A0A182JNG2 T1DIX5 A0A182VP76 A0A182UE48 Q7PM07 A0A182XCH4 A0A1I8PPX4 A0A182JH70 A0A182QT19 A0A182ICM9 A0A182F3S7 A0A1B0EX50 A0A0K8TPK0 U5EPQ1 D6X1A2 A0A1L8EEP8 V5GXL2 A0A1A9ZTH3 A0A1I8NGP2 A0A0A9ZHC0 A0A1A9UIT0 A0A1B0AKL7 A0A336K4K4 A0A0L0CN01 A0A1A9YA35 A0A1B0GD88 A0A1B6DKV6 A0A1W4WW49 B4KKX9 A0A226CVU9 B4N019 A0A224XZM4 A0A069DWA7 K7J429 A0A0A9WPN4 A0A2R7VSR6 B4M9J9 A0A1A9X493 B3ML22 A0A0V0G590 R4G4E8 J3JVU1 A0A3B0KFL7 A0A232EFC3 A0A0K8W7F0 B4JQ71 A0A1J1IJN1 W8CBZ4 A0A1W4V387 A0A034WT69 A0A0C9RNB9 B3N610 B4HXS7 B4Q6H4 A0A0M4E376 A0A023F7S0 A0A023F740 Q29KE7 Q9V3W2 B4HBZ0 B4NY88 A0A0A1X7J7 A0A0A1XM97 A0A1Y1M2M7 A0A293M241 A0A1D2MW98 R4FMV0 A0A1D2AI60 A0A0K8R9H0 B7QKY7
A0A2H1V7Z2 A0A2W1BRZ6 A0A2A4K4Z6 A0A0L7KNJ0 A0A1L8E162 A0A023EG32 A0A182H4X5 W5JUX8 A0A182MHR1 A0A2M4CV22 A0A2M3Z495 A0A2M4C1L4 T1DJ84 A0A2M4AFC2 A0A182WHI5 A0A182RRI5 A0A1Q3FIX2 A0A182NUW8 A0A2J7R2Q2 A0A182P3C2 A0A1B0D6Z1 B0W0C0 Q16WX6 A0A182Y5L0 A0A182JNG2 T1DIX5 A0A182VP76 A0A182UE48 Q7PM07 A0A182XCH4 A0A1I8PPX4 A0A182JH70 A0A182QT19 A0A182ICM9 A0A182F3S7 A0A1B0EX50 A0A0K8TPK0 U5EPQ1 D6X1A2 A0A1L8EEP8 V5GXL2 A0A1A9ZTH3 A0A1I8NGP2 A0A0A9ZHC0 A0A1A9UIT0 A0A1B0AKL7 A0A336K4K4 A0A0L0CN01 A0A1A9YA35 A0A1B0GD88 A0A1B6DKV6 A0A1W4WW49 B4KKX9 A0A226CVU9 B4N019 A0A224XZM4 A0A069DWA7 K7J429 A0A0A9WPN4 A0A2R7VSR6 B4M9J9 A0A1A9X493 B3ML22 A0A0V0G590 R4G4E8 J3JVU1 A0A3B0KFL7 A0A232EFC3 A0A0K8W7F0 B4JQ71 A0A1J1IJN1 W8CBZ4 A0A1W4V387 A0A034WT69 A0A0C9RNB9 B3N610 B4HXS7 B4Q6H4 A0A0M4E376 A0A023F7S0 A0A023F740 Q29KE7 Q9V3W2 B4HBZ0 B4NY88 A0A0A1X7J7 A0A0A1XM97 A0A1Y1M2M7 A0A293M241 A0A1D2MW98 R4FMV0 A0A1D2AI60 A0A0K8R9H0 B7QKY7
Pubmed
23622113
22118469
26354079
22651552
28756777
26227816
+ More
24945155 26483478 20920257 23761445 17510324 25244985 24330624 12364791 26369729 18362917 19820115 25315136 25401762 26823975 26108605 17994087 26334808 20075255 18057021 22516182 23537049 28648823 24495485 25348373 22936249 25474469 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 28004739 27289101
24945155 26483478 20920257 23761445 17510324 25244985 24330624 12364791 26369729 18362917 19820115 25315136 25401762 26823975 26108605 17994087 26334808 20075255 18057021 22516182 23537049 28648823 24495485 25348373 22936249 25474469 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 28004739 27289101
EMBL
DQ985604
ABJ97192.1
GAIX01013941
JAA78619.1
AGBW02008872
OWR52324.1
+ More
KQ460226 KPJ16462.1 AK401800 BAM18422.1 KQ459597 KPI95056.1 ODYU01001155 SOQ36968.1 KZ149992 PZC75560.1 NWSH01000167 PCG78843.1 JTDY01008281 KOB64645.1 GFDF01001849 JAV12235.1 GAPW01005235 JAC08363.1 JXUM01025859 JXUM01025860 KQ560736 KXJ81078.1 ADMH02000386 ETN66750.1 AXCM01005849 GGFL01004510 MBW68688.1 GGFM01002589 MBW23340.1 GGFJ01010062 MBW59203.1 GAMD01001591 JAA99999.1 GGFK01006159 MBW39480.1 GFDL01007508 JAV27537.1 NEVH01007828 PNF35118.1 AJVK01003818 AJVK01003819 DS231817 EDS39659.1 CH477551 EAT39112.1 GALA01000686 JAA94166.1 AAAB01008980 EAA14545.3 AXCN02000750 APCN01005735 AJWK01011153 AJWK01011154 GDAI01001319 JAI16284.1 GANO01004669 JAB55202.1 KQ971372 EFA10794.1 GFDG01001592 JAV17207.1 GALX01001944 JAB66522.1 GBHO01000481 GBRD01002986 GDHC01003181 JAG43123.1 JAG62835.1 JAQ15448.1 JXJN01026161 UFQS01000101 UFQT01000101 SSW99589.1 SSX19969.1 JRES01000175 KNC33592.1 CCAG010015652 GEDC01010972 JAS26326.1 CH933807 EDW11709.1 LNIX01000066 OXA37049.1 CH963920 EDW77954.1 GFTR01002703 JAW13723.1 GBGD01003280 JAC85609.1 AAZX01001009 GBHO01036794 JAG06810.1 KK854064 PTY10553.1 CH940654 EDW57875.1 KRF77996.1 CH902620 EDV30680.1 KPU72988.1 GECL01002913 JAP03211.1 GAHY01001425 JAA76085.1 APGK01050136 BT127359 KB741166 KB632257 AEE62321.1 ENN73379.1 ERL90777.1 OUUW01000006 SPP82428.1 NNAY01005063 OXU17051.1 GDHF01009374 GDHF01005539 JAI42940.1 JAI46775.1 CH916372 EDV99051.1 CVRI01000054 CRL00455.1 GAMC01000089 GAMC01000088 JAC06467.1 GAKP01000241 JAC58711.1 GBYB01008576 JAG78343.1 CH954177 EDV58048.1 CH480818 EDW51857.1 CM000361 CM002910 EDX05156.1 KMY90402.1 CP012523 ALC39042.1 ALC40216.1 GBBI01001425 JAC17287.1 GBBI01001657 JAC17055.1 CH379061 EAL33228.2 AE014134 BT001463 AAF53480.1 AAN10909.2 AAN71218.1 CH479272 EDW40001.1 CM000157 EDW89724.1 GBXI01007456 JAD06836.1 GBXI01002599 JAD11693.1 GEZM01041912 GEZM01041911 JAV80102.1 GFWV01010314 MAA35043.1 LJIJ01000461 ODM97228.1 ACPB03028724 GAHY01001584 JAA75926.1 GETE01000866 JAT78880.1 GADI01006087 JAA67721.1 ABJB010660488 DS962636 EEC19509.1
KQ460226 KPJ16462.1 AK401800 BAM18422.1 KQ459597 KPI95056.1 ODYU01001155 SOQ36968.1 KZ149992 PZC75560.1 NWSH01000167 PCG78843.1 JTDY01008281 KOB64645.1 GFDF01001849 JAV12235.1 GAPW01005235 JAC08363.1 JXUM01025859 JXUM01025860 KQ560736 KXJ81078.1 ADMH02000386 ETN66750.1 AXCM01005849 GGFL01004510 MBW68688.1 GGFM01002589 MBW23340.1 GGFJ01010062 MBW59203.1 GAMD01001591 JAA99999.1 GGFK01006159 MBW39480.1 GFDL01007508 JAV27537.1 NEVH01007828 PNF35118.1 AJVK01003818 AJVK01003819 DS231817 EDS39659.1 CH477551 EAT39112.1 GALA01000686 JAA94166.1 AAAB01008980 EAA14545.3 AXCN02000750 APCN01005735 AJWK01011153 AJWK01011154 GDAI01001319 JAI16284.1 GANO01004669 JAB55202.1 KQ971372 EFA10794.1 GFDG01001592 JAV17207.1 GALX01001944 JAB66522.1 GBHO01000481 GBRD01002986 GDHC01003181 JAG43123.1 JAG62835.1 JAQ15448.1 JXJN01026161 UFQS01000101 UFQT01000101 SSW99589.1 SSX19969.1 JRES01000175 KNC33592.1 CCAG010015652 GEDC01010972 JAS26326.1 CH933807 EDW11709.1 LNIX01000066 OXA37049.1 CH963920 EDW77954.1 GFTR01002703 JAW13723.1 GBGD01003280 JAC85609.1 AAZX01001009 GBHO01036794 JAG06810.1 KK854064 PTY10553.1 CH940654 EDW57875.1 KRF77996.1 CH902620 EDV30680.1 KPU72988.1 GECL01002913 JAP03211.1 GAHY01001425 JAA76085.1 APGK01050136 BT127359 KB741166 KB632257 AEE62321.1 ENN73379.1 ERL90777.1 OUUW01000006 SPP82428.1 NNAY01005063 OXU17051.1 GDHF01009374 GDHF01005539 JAI42940.1 JAI46775.1 CH916372 EDV99051.1 CVRI01000054 CRL00455.1 GAMC01000089 GAMC01000088 JAC06467.1 GAKP01000241 JAC58711.1 GBYB01008576 JAG78343.1 CH954177 EDV58048.1 CH480818 EDW51857.1 CM000361 CM002910 EDX05156.1 KMY90402.1 CP012523 ALC39042.1 ALC40216.1 GBBI01001425 JAC17287.1 GBBI01001657 JAC17055.1 CH379061 EAL33228.2 AE014134 BT001463 AAF53480.1 AAN10909.2 AAN71218.1 CH479272 EDW40001.1 CM000157 EDW89724.1 GBXI01007456 JAD06836.1 GBXI01002599 JAD11693.1 GEZM01041912 GEZM01041911 JAV80102.1 GFWV01010314 MAA35043.1 LJIJ01000461 ODM97228.1 ACPB03028724 GAHY01001584 JAA75926.1 GETE01000866 JAT78880.1 GADI01006087 JAA67721.1 ABJB010660488 DS962636 EEC19509.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000218220
UP000037510
UP000069940
+ More
UP000249989 UP000000673 UP000075883 UP000075920 UP000075900 UP000075884 UP000235965 UP000075885 UP000092462 UP000002320 UP000008820 UP000076408 UP000075881 UP000075903 UP000075902 UP000007062 UP000076407 UP000095300 UP000075880 UP000075886 UP000075840 UP000069272 UP000092461 UP000007266 UP000092445 UP000095301 UP000078200 UP000092460 UP000037069 UP000092443 UP000092444 UP000192223 UP000009192 UP000198287 UP000007798 UP000002358 UP000008792 UP000091820 UP000007801 UP000019118 UP000030742 UP000268350 UP000215335 UP000001070 UP000183832 UP000192221 UP000008711 UP000001292 UP000000304 UP000092553 UP000001819 UP000000803 UP000008744 UP000002282 UP000094527 UP000015103 UP000001555
UP000249989 UP000000673 UP000075883 UP000075920 UP000075900 UP000075884 UP000235965 UP000075885 UP000092462 UP000002320 UP000008820 UP000076408 UP000075881 UP000075903 UP000075902 UP000007062 UP000076407 UP000095300 UP000075880 UP000075886 UP000075840 UP000069272 UP000092461 UP000007266 UP000092445 UP000095301 UP000078200 UP000092460 UP000037069 UP000092443 UP000092444 UP000192223 UP000009192 UP000198287 UP000007798 UP000002358 UP000008792 UP000091820 UP000007801 UP000019118 UP000030742 UP000268350 UP000215335 UP000001070 UP000183832 UP000192221 UP000008711 UP000001292 UP000000304 UP000092553 UP000001819 UP000000803 UP000008744 UP000002282 UP000094527 UP000015103 UP000001555
PRIDE
Interpro
ProteinModelPortal
A0FDR2
S4NNG4
A0A212FF15
A0A0N1IPM2
I4DKI2
A0A194PNP8
+ More
A0A2H1V7Z2 A0A2W1BRZ6 A0A2A4K4Z6 A0A0L7KNJ0 A0A1L8E162 A0A023EG32 A0A182H4X5 W5JUX8 A0A182MHR1 A0A2M4CV22 A0A2M3Z495 A0A2M4C1L4 T1DJ84 A0A2M4AFC2 A0A182WHI5 A0A182RRI5 A0A1Q3FIX2 A0A182NUW8 A0A2J7R2Q2 A0A182P3C2 A0A1B0D6Z1 B0W0C0 Q16WX6 A0A182Y5L0 A0A182JNG2 T1DIX5 A0A182VP76 A0A182UE48 Q7PM07 A0A182XCH4 A0A1I8PPX4 A0A182JH70 A0A182QT19 A0A182ICM9 A0A182F3S7 A0A1B0EX50 A0A0K8TPK0 U5EPQ1 D6X1A2 A0A1L8EEP8 V5GXL2 A0A1A9ZTH3 A0A1I8NGP2 A0A0A9ZHC0 A0A1A9UIT0 A0A1B0AKL7 A0A336K4K4 A0A0L0CN01 A0A1A9YA35 A0A1B0GD88 A0A1B6DKV6 A0A1W4WW49 B4KKX9 A0A226CVU9 B4N019 A0A224XZM4 A0A069DWA7 K7J429 A0A0A9WPN4 A0A2R7VSR6 B4M9J9 A0A1A9X493 B3ML22 A0A0V0G590 R4G4E8 J3JVU1 A0A3B0KFL7 A0A232EFC3 A0A0K8W7F0 B4JQ71 A0A1J1IJN1 W8CBZ4 A0A1W4V387 A0A034WT69 A0A0C9RNB9 B3N610 B4HXS7 B4Q6H4 A0A0M4E376 A0A023F7S0 A0A023F740 Q29KE7 Q9V3W2 B4HBZ0 B4NY88 A0A0A1X7J7 A0A0A1XM97 A0A1Y1M2M7 A0A293M241 A0A1D2MW98 R4FMV0 A0A1D2AI60 A0A0K8R9H0 B7QKY7
A0A2H1V7Z2 A0A2W1BRZ6 A0A2A4K4Z6 A0A0L7KNJ0 A0A1L8E162 A0A023EG32 A0A182H4X5 W5JUX8 A0A182MHR1 A0A2M4CV22 A0A2M3Z495 A0A2M4C1L4 T1DJ84 A0A2M4AFC2 A0A182WHI5 A0A182RRI5 A0A1Q3FIX2 A0A182NUW8 A0A2J7R2Q2 A0A182P3C2 A0A1B0D6Z1 B0W0C0 Q16WX6 A0A182Y5L0 A0A182JNG2 T1DIX5 A0A182VP76 A0A182UE48 Q7PM07 A0A182XCH4 A0A1I8PPX4 A0A182JH70 A0A182QT19 A0A182ICM9 A0A182F3S7 A0A1B0EX50 A0A0K8TPK0 U5EPQ1 D6X1A2 A0A1L8EEP8 V5GXL2 A0A1A9ZTH3 A0A1I8NGP2 A0A0A9ZHC0 A0A1A9UIT0 A0A1B0AKL7 A0A336K4K4 A0A0L0CN01 A0A1A9YA35 A0A1B0GD88 A0A1B6DKV6 A0A1W4WW49 B4KKX9 A0A226CVU9 B4N019 A0A224XZM4 A0A069DWA7 K7J429 A0A0A9WPN4 A0A2R7VSR6 B4M9J9 A0A1A9X493 B3ML22 A0A0V0G590 R4G4E8 J3JVU1 A0A3B0KFL7 A0A232EFC3 A0A0K8W7F0 B4JQ71 A0A1J1IJN1 W8CBZ4 A0A1W4V387 A0A034WT69 A0A0C9RNB9 B3N610 B4HXS7 B4Q6H4 A0A0M4E376 A0A023F7S0 A0A023F740 Q29KE7 Q9V3W2 B4HBZ0 B4NY88 A0A0A1X7J7 A0A0A1XM97 A0A1Y1M2M7 A0A293M241 A0A1D2MW98 R4FMV0 A0A1D2AI60 A0A0K8R9H0 B7QKY7
PDB
5LNK
E-value=0.0271263,
Score=82
Ontologies
GO
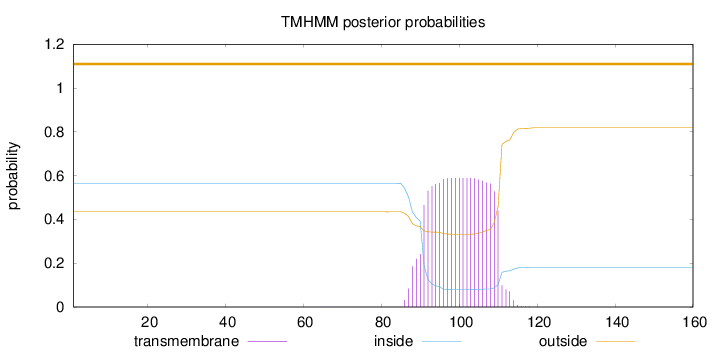
Topology
Length:
160
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.28994
Exp number, first 60 AAs:
0
Total prob of N-in:
0.56535
outside
1 - 160
Population Genetic Test Statistics
Pi
128.576367
Theta
114.256506
Tajima's D
0.157699
CLR
0.667788
CSRT
0.414979251037448
Interpretation
Uncertain
