Gene
KWMTBOMO02935
Annotation
PREDICTED:_probable_dimethyladenosine_transferase_[Amyelois_transitella]
Full name
rRNA adenine N(6)-methyltransferase
Location in the cell
Cytoplasmic Reliability : 1.465 Mitochondrial Reliability : 1.443 Nuclear Reliability : 1.365
Sequence
CDS
ATGCCAAAAATTAAGGCAGAAAAAAAAACGAGAATTCATAAAGAAATTGCTAAACAAGGTATTCAATTTAACAAGGACTTCGGTCAACACATATTGAAGAATCCACTAATAATAACCTCTATGTTAGACAAATCTGGATTAAGGCCTACTGATGTAGCACTTGAGATTGGTCCTGGTACTGGCAATATGACAGTCAAATTGCTGGACAGAGTAAAAAAGGTATTAGCCTGTGAAATTGATACAAGATTGGTTGCTGAACTTCAAAAACGTGTGCAAGGTACTCCTTATCAAGCAAAATTACAAATTTTGGTTGGAGATGTTTTGAAAACAGAATTACCATTTTTTGATATATGTGTAGCAAATATTCCATATCAAATTAGTTCACCTTTAGTGTTCAAATTGCTGCTACACAGACCTTTCTTTAGGTGTGCAGTGCTCATGTTTCAAAAGGAGTTTGCACAGAGATTAGTTGCAAAGCCTGGTGATAAATTGTACTGCAGACTGTCAATAAATACACAATTATTGGCTAGAGTTGATATGCTGATGAAGGTTGGCAAGAACAATTTCAGACCACCACCCAAAGTAGAGTCTAGTGTTGTGAGAATAGAACCAAGAAATCCACCTCCGCCTATAAATTTTGTAGAATGGGATGGTTTGACTAGAATAGCATTTGTAAGAAAAAATAAAACACTCTCAGCTGCATTCAAACATGCAACCACAATGGCTGCTTTGGAGAAAAATTACAGGGTCCATTGTTCTTTACACAATAAGGAAATCCCTGAAGATTTTGATATTAAGCAAAAAGTACATGAAATATTGACATCCTGTGAAGCTGATCAAATGAGGGCTAGAACAATGGATATTGATGACTTTATGAAGCTATTACATGCTTTTAATGCTGAAGGGATTCATTTTGCATAA
Protein
MPKIKAEKKTRIHKEIAKQGIQFNKDFGQHILKNPLIITSMLDKSGLRPTDVALEIGPGTGNMTVKLLDRVKKVLACEIDTRLVAELQKRVQGTPYQAKLQILVGDVLKTELPFFDICVANIPYQISSPLVFKLLLHRPFFRCAVLMFQKEFAQRLVAKPGDKLYCRLSINTQLLARVDMLMKVGKNNFRPPPKVESSVVRIEPRNPPPPINFVEWDGLTRIAFVRKNKTLSAAFKHATTMAALEKNYRVHCSLHNKEIPEDFDIKQKVHEILTSCEADQMRARTMDIDDFMKLLHAFNAEGIHFA
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family.
Uniprot
A0A212FEZ4
A0A2W1BKJ4
A0A0N0PDE6
A0A194PV98
A0A2A4K589
A0A2H1V834
+ More
A0A1B0DGY8 A0A067RF98 A0A2P8XP70 A0A1B6M3C2 A0A1L8DGW9 A0A2J7QNZ0 B0X533 A0A1Y1N263 A0A1Q3F394 Q16SG3 A0A023EQC4 S4NNU2 A0A154P3E6 J3JYM3 A0A336MJK0 A0A084VDD6 A0A1B6IVQ6 A0A2M4AUZ7 A0A2M4BV62 A0A1B6GX66 A0A182PTB9 A0A182J7L4 A0A182FSE4 A0A1B3PDB2 A0A2M4BV44 W5J7T1 D6X457 A0A182QRT1 A0A182Y8V0 A0A0C9R4Z0 A0A2M3ZC60 A0A1B6CFY7 A0A2M3ZBD2 A0A2M4AV57 A0A182TJ86 A0A182KZM6 A0A182XFC0 Q7QA26 A0A182HYL2 A0A2M3ZBG3 A0A0M9A8S6 A0A1B3PDI5 A0A182LWA9 A0A1J1I9L5 A0A182RT49 T1PJM4 A0A1I8MLW2 A0A3B4CS58 A0A2D0R005 A0A158NMJ9 A0A151I3E2 E2B908 A0A195C3P3 A0A0L0CKB2 A0A034VAS2 W8B474 B0S688 I3K854 A0A2A3E7P8 F1QJ20 A0A0K8WC26 A0A3B5BKU5 K7J3U2 A0A3P8ZUI8 A0A232F4Z5 A0A1I8P6S0 A0A222AJ23 A0A151K0R3 A0A3B3BGI3 A0A3P8TNT9 A0A3B4GR01 A0A3P8NLY0 Q298Z5 B4G4X1 A0A222AJE0 A0A1S3IQZ2 W5MMA5 C1BZP0 A0A0S7LSB9 A0A3B3XQ33 M3ZY81 A0A087YIK9 A0A3B3VVV4 Q6DC09 A0A3P9CL52 H2TCE0 A0A3B0K1A9 A0A060YLS9 A0A1A7XQI7 A0A2U9BRX0 B3LXN7 A0A3B4WJA0 A0A3B4USJ3 C3YM50
A0A1B0DGY8 A0A067RF98 A0A2P8XP70 A0A1B6M3C2 A0A1L8DGW9 A0A2J7QNZ0 B0X533 A0A1Y1N263 A0A1Q3F394 Q16SG3 A0A023EQC4 S4NNU2 A0A154P3E6 J3JYM3 A0A336MJK0 A0A084VDD6 A0A1B6IVQ6 A0A2M4AUZ7 A0A2M4BV62 A0A1B6GX66 A0A182PTB9 A0A182J7L4 A0A182FSE4 A0A1B3PDB2 A0A2M4BV44 W5J7T1 D6X457 A0A182QRT1 A0A182Y8V0 A0A0C9R4Z0 A0A2M3ZC60 A0A1B6CFY7 A0A2M3ZBD2 A0A2M4AV57 A0A182TJ86 A0A182KZM6 A0A182XFC0 Q7QA26 A0A182HYL2 A0A2M3ZBG3 A0A0M9A8S6 A0A1B3PDI5 A0A182LWA9 A0A1J1I9L5 A0A182RT49 T1PJM4 A0A1I8MLW2 A0A3B4CS58 A0A2D0R005 A0A158NMJ9 A0A151I3E2 E2B908 A0A195C3P3 A0A0L0CKB2 A0A034VAS2 W8B474 B0S688 I3K854 A0A2A3E7P8 F1QJ20 A0A0K8WC26 A0A3B5BKU5 K7J3U2 A0A3P8ZUI8 A0A232F4Z5 A0A1I8P6S0 A0A222AJ23 A0A151K0R3 A0A3B3BGI3 A0A3P8TNT9 A0A3B4GR01 A0A3P8NLY0 Q298Z5 B4G4X1 A0A222AJE0 A0A1S3IQZ2 W5MMA5 C1BZP0 A0A0S7LSB9 A0A3B3XQ33 M3ZY81 A0A087YIK9 A0A3B3VVV4 Q6DC09 A0A3P9CL52 H2TCE0 A0A3B0K1A9 A0A060YLS9 A0A1A7XQI7 A0A2U9BRX0 B3LXN7 A0A3B4WJA0 A0A3B4USJ3 C3YM50
EC Number
2.1.1.-
Pubmed
22118469
28756777
26354079
24845553
29403074
28004739
+ More
17510324 24945155 23622113 22516182 23537049 24438588 20920257 23761445 18362917 19820115 25244985 20966253 12364791 14747013 17210077 25315136 21347285 20798317 26108605 25348373 24495485 23594743 25186727 20075255 25069045 28648823 29451363 15632085 17994087 20433749 23542700 21551351 24755649 18563158
17510324 24945155 23622113 22516182 23537049 24438588 20920257 23761445 18362917 19820115 25244985 20966253 12364791 14747013 17210077 25315136 21347285 20798317 26108605 25348373 24495485 23594743 25186727 20075255 25069045 28648823 29451363 15632085 17994087 20433749 23542700 21551351 24755649 18563158
EMBL
AGBW02008872
OWR52325.1
KZ149992
PZC75559.1
KQ460226
KPJ16461.1
+ More
KQ459597 KPI95055.1 NWSH01000167 PCG78842.1 ODYU01001155 SOQ36969.1 AJVK01029492 AJVK01033907 KK852543 KDR21683.1 PYGN01001603 PSN33798.1 GEBQ01009555 JAT30422.1 GFDF01008385 JAV05699.1 NEVH01012098 PNF30301.1 DS232362 EDS40679.1 GEZM01017357 JAV90755.1 GFDL01013013 JAV22032.1 CH477673 CH477186 EAT37437.1 EAT48949.1 GAPW01002834 JAC10764.1 GAIX01012139 JAA80421.1 KQ434796 KZC05758.1 APGK01028302 BT128352 KB740686 KB631930 AEE63310.1 ENN79543.1 ERL87302.1 UFQT01001439 SSX30582.1 ATLV01011260 KE524659 KFB35980.1 GECU01016680 JAS91026.1 GGFK01011270 MBW44591.1 GGFJ01007741 MBW56882.1 GECZ01002743 JAS67026.1 KU659918 AOG17717.1 GGFJ01007742 MBW56883.1 ADMH02001999 ETN60036.1 KQ971379 EEZ97508.1 AXCN02000234 GBYB01007832 JAG77599.1 GGFM01005287 MBW26038.1 GEDC01025043 JAS12255.1 GGFM01005103 MBW25854.1 GGFK01011271 MBW44592.1 AAAB01008898 EAA09266.2 APCN01005389 GGFM01005143 MBW25894.1 KQ435711 KOX79550.1 KU659994 AOG17792.1 AXCM01000209 CVRI01000045 CRK96936.1 KA648305 AFP62934.1 ADTU01002284 ADTU01002285 KQ976483 KYM83698.1 GL446405 EFN87842.1 KQ978344 KYM94806.1 JRES01000370 KNC31889.1 GAKP01019340 GAKP01019338 JAC39614.1 GAMC01013178 GAMC01013176 JAB93377.1 BX465215 AERX01026878 KZ288340 PBC27783.1 GDHF01026986 GDHF01022583 GDHF01013500 GDHF01003705 JAI25328.1 JAI29731.1 JAI38814.1 JAI48609.1 NNAY01000925 OXU25874.1 KY563977 ASO76385.1 KQ981253 KYN44197.1 CM000070 EAL27808.1 CH479179 EDW24637.1 KY609807 ASO76496.1 AHAT01002686 BT080069 ACO14493.1 GBYX01152068 JAO92314.1 AYCK01007762 BC078286 AAH78286.1 OUUW01000013 SPP88057.1 FR910148 CDQ90095.1 HADW01018709 HADX01009900 SBP20109.1 CP026251 AWP06967.1 CH902617 EDV43931.1 GG666529 EEN58644.1
KQ459597 KPI95055.1 NWSH01000167 PCG78842.1 ODYU01001155 SOQ36969.1 AJVK01029492 AJVK01033907 KK852543 KDR21683.1 PYGN01001603 PSN33798.1 GEBQ01009555 JAT30422.1 GFDF01008385 JAV05699.1 NEVH01012098 PNF30301.1 DS232362 EDS40679.1 GEZM01017357 JAV90755.1 GFDL01013013 JAV22032.1 CH477673 CH477186 EAT37437.1 EAT48949.1 GAPW01002834 JAC10764.1 GAIX01012139 JAA80421.1 KQ434796 KZC05758.1 APGK01028302 BT128352 KB740686 KB631930 AEE63310.1 ENN79543.1 ERL87302.1 UFQT01001439 SSX30582.1 ATLV01011260 KE524659 KFB35980.1 GECU01016680 JAS91026.1 GGFK01011270 MBW44591.1 GGFJ01007741 MBW56882.1 GECZ01002743 JAS67026.1 KU659918 AOG17717.1 GGFJ01007742 MBW56883.1 ADMH02001999 ETN60036.1 KQ971379 EEZ97508.1 AXCN02000234 GBYB01007832 JAG77599.1 GGFM01005287 MBW26038.1 GEDC01025043 JAS12255.1 GGFM01005103 MBW25854.1 GGFK01011271 MBW44592.1 AAAB01008898 EAA09266.2 APCN01005389 GGFM01005143 MBW25894.1 KQ435711 KOX79550.1 KU659994 AOG17792.1 AXCM01000209 CVRI01000045 CRK96936.1 KA648305 AFP62934.1 ADTU01002284 ADTU01002285 KQ976483 KYM83698.1 GL446405 EFN87842.1 KQ978344 KYM94806.1 JRES01000370 KNC31889.1 GAKP01019340 GAKP01019338 JAC39614.1 GAMC01013178 GAMC01013176 JAB93377.1 BX465215 AERX01026878 KZ288340 PBC27783.1 GDHF01026986 GDHF01022583 GDHF01013500 GDHF01003705 JAI25328.1 JAI29731.1 JAI38814.1 JAI48609.1 NNAY01000925 OXU25874.1 KY563977 ASO76385.1 KQ981253 KYN44197.1 CM000070 EAL27808.1 CH479179 EDW24637.1 KY609807 ASO76496.1 AHAT01002686 BT080069 ACO14493.1 GBYX01152068 JAO92314.1 AYCK01007762 BC078286 AAH78286.1 OUUW01000013 SPP88057.1 FR910148 CDQ90095.1 HADW01018709 HADX01009900 SBP20109.1 CP026251 AWP06967.1 CH902617 EDV43931.1 GG666529 EEN58644.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000218220
UP000092462
UP000027135
+ More
UP000245037 UP000235965 UP000002320 UP000008820 UP000076502 UP000019118 UP000030742 UP000030765 UP000075885 UP000075880 UP000069272 UP000000673 UP000007266 UP000075886 UP000076408 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000053105 UP000075883 UP000183832 UP000075900 UP000095301 UP000261440 UP000221080 UP000005205 UP000078540 UP000008237 UP000078542 UP000037069 UP000000437 UP000005207 UP000242457 UP000261400 UP000002358 UP000265140 UP000215335 UP000095300 UP000078541 UP000261560 UP000265080 UP000261460 UP000265100 UP000001819 UP000008744 UP000085678 UP000018468 UP000261480 UP000002852 UP000028760 UP000261500 UP000265160 UP000005226 UP000268350 UP000193380 UP000246464 UP000007801 UP000261360 UP000261420 UP000001554
UP000245037 UP000235965 UP000002320 UP000008820 UP000076502 UP000019118 UP000030742 UP000030765 UP000075885 UP000075880 UP000069272 UP000000673 UP000007266 UP000075886 UP000076408 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000053105 UP000075883 UP000183832 UP000075900 UP000095301 UP000261440 UP000221080 UP000005205 UP000078540 UP000008237 UP000078542 UP000037069 UP000000437 UP000005207 UP000242457 UP000261400 UP000002358 UP000265140 UP000215335 UP000095300 UP000078541 UP000261560 UP000265080 UP000261460 UP000265100 UP000001819 UP000008744 UP000085678 UP000018468 UP000261480 UP000002852 UP000028760 UP000261500 UP000265160 UP000005226 UP000268350 UP000193380 UP000246464 UP000007801 UP000261360 UP000261420 UP000001554
Pfam
PF00398 RrnaAD
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
A0A212FEZ4
A0A2W1BKJ4
A0A0N0PDE6
A0A194PV98
A0A2A4K589
A0A2H1V834
+ More
A0A1B0DGY8 A0A067RF98 A0A2P8XP70 A0A1B6M3C2 A0A1L8DGW9 A0A2J7QNZ0 B0X533 A0A1Y1N263 A0A1Q3F394 Q16SG3 A0A023EQC4 S4NNU2 A0A154P3E6 J3JYM3 A0A336MJK0 A0A084VDD6 A0A1B6IVQ6 A0A2M4AUZ7 A0A2M4BV62 A0A1B6GX66 A0A182PTB9 A0A182J7L4 A0A182FSE4 A0A1B3PDB2 A0A2M4BV44 W5J7T1 D6X457 A0A182QRT1 A0A182Y8V0 A0A0C9R4Z0 A0A2M3ZC60 A0A1B6CFY7 A0A2M3ZBD2 A0A2M4AV57 A0A182TJ86 A0A182KZM6 A0A182XFC0 Q7QA26 A0A182HYL2 A0A2M3ZBG3 A0A0M9A8S6 A0A1B3PDI5 A0A182LWA9 A0A1J1I9L5 A0A182RT49 T1PJM4 A0A1I8MLW2 A0A3B4CS58 A0A2D0R005 A0A158NMJ9 A0A151I3E2 E2B908 A0A195C3P3 A0A0L0CKB2 A0A034VAS2 W8B474 B0S688 I3K854 A0A2A3E7P8 F1QJ20 A0A0K8WC26 A0A3B5BKU5 K7J3U2 A0A3P8ZUI8 A0A232F4Z5 A0A1I8P6S0 A0A222AJ23 A0A151K0R3 A0A3B3BGI3 A0A3P8TNT9 A0A3B4GR01 A0A3P8NLY0 Q298Z5 B4G4X1 A0A222AJE0 A0A1S3IQZ2 W5MMA5 C1BZP0 A0A0S7LSB9 A0A3B3XQ33 M3ZY81 A0A087YIK9 A0A3B3VVV4 Q6DC09 A0A3P9CL52 H2TCE0 A0A3B0K1A9 A0A060YLS9 A0A1A7XQI7 A0A2U9BRX0 B3LXN7 A0A3B4WJA0 A0A3B4USJ3 C3YM50
A0A1B0DGY8 A0A067RF98 A0A2P8XP70 A0A1B6M3C2 A0A1L8DGW9 A0A2J7QNZ0 B0X533 A0A1Y1N263 A0A1Q3F394 Q16SG3 A0A023EQC4 S4NNU2 A0A154P3E6 J3JYM3 A0A336MJK0 A0A084VDD6 A0A1B6IVQ6 A0A2M4AUZ7 A0A2M4BV62 A0A1B6GX66 A0A182PTB9 A0A182J7L4 A0A182FSE4 A0A1B3PDB2 A0A2M4BV44 W5J7T1 D6X457 A0A182QRT1 A0A182Y8V0 A0A0C9R4Z0 A0A2M3ZC60 A0A1B6CFY7 A0A2M3ZBD2 A0A2M4AV57 A0A182TJ86 A0A182KZM6 A0A182XFC0 Q7QA26 A0A182HYL2 A0A2M3ZBG3 A0A0M9A8S6 A0A1B3PDI5 A0A182LWA9 A0A1J1I9L5 A0A182RT49 T1PJM4 A0A1I8MLW2 A0A3B4CS58 A0A2D0R005 A0A158NMJ9 A0A151I3E2 E2B908 A0A195C3P3 A0A0L0CKB2 A0A034VAS2 W8B474 B0S688 I3K854 A0A2A3E7P8 F1QJ20 A0A0K8WC26 A0A3B5BKU5 K7J3U2 A0A3P8ZUI8 A0A232F4Z5 A0A1I8P6S0 A0A222AJ23 A0A151K0R3 A0A3B3BGI3 A0A3P8TNT9 A0A3B4GR01 A0A3P8NLY0 Q298Z5 B4G4X1 A0A222AJE0 A0A1S3IQZ2 W5MMA5 C1BZP0 A0A0S7LSB9 A0A3B3XQ33 M3ZY81 A0A087YIK9 A0A3B3VVV4 Q6DC09 A0A3P9CL52 H2TCE0 A0A3B0K1A9 A0A060YLS9 A0A1A7XQI7 A0A2U9BRX0 B3LXN7 A0A3B4WJA0 A0A3B4USJ3 C3YM50
PDB
1ZQ9
E-value=3.67765e-120,
Score=1103
Ontologies
KEGG
GO
PANTHER
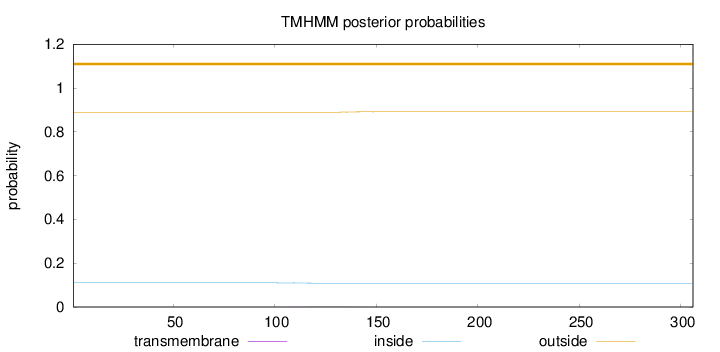
Topology
Length:
306
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0535900000000001
Exp number, first 60 AAs:
0.000740000000000001
Total prob of N-in:
0.11026
outside
1 - 306
Population Genetic Test Statistics
Pi
148.030095
Theta
143.90861
Tajima's D
0.186901
CLR
0.531502
CSRT
0.419979001049948
Interpretation
Uncertain
