Gene
KWMTBOMO02924
Pre Gene Modal
BGIBMGA003552
Annotation
Protein_rolling_stone_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.793
Sequence
CDS
ATGAACGACAAATTCGGTTACTGGTTCATTTATCTCACACATTGGGGCCTCACGATCAACACTCTGGCGACGGGTTTTGCAACTGCTGTTTCTGCACGTTGCTACTTCTACGGACCAATTAGCGCTGAATTCAAACTACCGTGGTATGTCAAGGTTTCCTGGGTACTCTACAACATTGCCGTACCACTCGCATTCCTGATCACAATTTTCTATTGGTCTTTACTTTTTGAGGCTGGAATTGAAGAAGAGTTAAACCACGCCTTGGATGTGGCCATCCACGGTATCAACTCCTTGGTAATGTTCATGTTGATGTCGAGCTCTTTGCACCCAAGTCGCTTACTACACATTTATCAACCCTTAACCTTCTCCGTGGTTTATGTCACCTTCGGGATTATATACTACTTCGCTGGTGGGGTTGAACAAGAAGGAAACCCTTGGATTTATCCGGTAGTGAATTGGGCCAACCCTGGACCCACTGTCGGGGTTGTGGCTTTGACTGGACTACTTCTGGTCATATTGCACTCAGTAACAGTGGCGTTTTCCGTTGGCCGAGACAAACTCTCTAAAAAGTTCCTTAATGCCACATCACCAAACGTACCTGAGGAGGTGCCTCTGCGGCAGCCTACACTAACTCCAACAGCAACAGTACAACTAAATGACAGCTGA
Protein
MNDKFGYWFIYLTHWGLTINTLATGFATAVSARCYFYGPISAEFKLPWYVKVSWVLYNIAVPLAFLITIFYWSLLFEAGIEEELNHALDVAIHGINSLVMFMLMSSSLHPSRLLHIYQPLTFSVVYVTFGIIYYFAGGVEQEGNPWIYPVVNWANPGPTVGVVALTGLLLVILHSVTVAFSVGRDKLSKKFLNATSPNVPEEVPLRQPTLTPTATVQLNDS
Summary
Uniprot
H9J215
A0A194PPZ8
A0A2A4JU86
A0A0N0PDH4
A0A1E1WH93
A0A212EPX0
+ More
A0A2A4JUA0 A0A0L7LFH1 A0A2H1V2M7 A0A0N1PGW6 A0A2A4JVE8 A0A1E1WUI9 A0A2W1BUW2 A0A2W1BMM1 A0A194PP05 A0A2W1BKD6 A0A0N1IPM0 A0A2A4JV90 A0A2H1V2I5 A0A2A4JVT0 A0A0N0PDS5 A0A194PQM7 A0A2A4JU90 A0A212EPW8 A0A212EPY3 A0A2W1BPQ3 A0A182QXJ3 A0A182F236 A0A2H1VVB3 A0A182Y5M5 A0A182IPP2 A0A1Q3FL99 A0A1Y1N4E4 A0A1Y1N4B9 A0A182MSD6 A0A182UZG0 A0A1Y9GLK9 A0A182L5X9 A0A1W4WQF2 A0A182RRG8 B0WX62 H9J216 A0A0M4EQI5 A0A1B6C3D6 A0A182GNM4 A0A336MIU4 B4NXK9 Q16S04 D6W7E7 B4HYT5 B3N7V9 B4Q7C1 A0A182XCJ4 B3MUQ8 Q9VLG1 A0A1B0CZ86 A0A1W4VZN2 B4MUK6 A0A1B6JNH2 B4LUZ2 Q29JS3 A0A0R3P206 A0A1L8D9A3 A0A3B0JUR8 B4GJ84 A0A1L8D935 S4PPR9 A0A1I8NB47 B4JZD8 A0A182WHK1 A0A2W1BKH1 W8B2M2 U4UNE7 N6U5I2 B4KFB2 W5JPB6 A0A0A1WH74 A0A182PB14
A0A2A4JUA0 A0A0L7LFH1 A0A2H1V2M7 A0A0N1PGW6 A0A2A4JVE8 A0A1E1WUI9 A0A2W1BUW2 A0A2W1BMM1 A0A194PP05 A0A2W1BKD6 A0A0N1IPM0 A0A2A4JV90 A0A2H1V2I5 A0A2A4JVT0 A0A0N0PDS5 A0A194PQM7 A0A2A4JU90 A0A212EPW8 A0A212EPY3 A0A2W1BPQ3 A0A182QXJ3 A0A182F236 A0A2H1VVB3 A0A182Y5M5 A0A182IPP2 A0A1Q3FL99 A0A1Y1N4E4 A0A1Y1N4B9 A0A182MSD6 A0A182UZG0 A0A1Y9GLK9 A0A182L5X9 A0A1W4WQF2 A0A182RRG8 B0WX62 H9J216 A0A0M4EQI5 A0A1B6C3D6 A0A182GNM4 A0A336MIU4 B4NXK9 Q16S04 D6W7E7 B4HYT5 B3N7V9 B4Q7C1 A0A182XCJ4 B3MUQ8 Q9VLG1 A0A1B0CZ86 A0A1W4VZN2 B4MUK6 A0A1B6JNH2 B4LUZ2 Q29JS3 A0A0R3P206 A0A1L8D9A3 A0A3B0JUR8 B4GJ84 A0A1L8D935 S4PPR9 A0A1I8NB47 B4JZD8 A0A182WHK1 A0A2W1BKH1 W8B2M2 U4UNE7 N6U5I2 B4KFB2 W5JPB6 A0A0A1WH74 A0A182PB14
Pubmed
19121390
26354079
22118469
26227816
28756777
25244985
+ More
28004739 20966253 26483478 17994087 17550304 17510324 18362917 19820115 18057021 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23622113 25315136 24495485 23537049 20920257 23761445 25830018
28004739 20966253 26483478 17994087 17550304 17510324 18362917 19820115 18057021 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23622113 25315136 24495485 23537049 20920257 23761445 25830018
EMBL
BABH01007552
BABH01007553
KQ459597
KPI95043.1
NWSH01000568
PCG75595.1
+ More
KQ460226 KPJ16448.1 GDQN01004690 JAT86364.1 AGBW02013358 OWR43540.1 PCG75607.1 JTDY01001287 KOB74293.1 ODYU01000378 SOQ35059.1 KPJ16447.1 PCG75594.1 GDQN01000412 JAT90642.1 KZ149992 PZC75543.1 PZC75541.1 KPI95042.1 PZC75542.1 KPJ16446.1 PCG75608.1 SOQ35058.1 PCG75593.1 KPJ16449.1 KPI95044.1 PCG75597.1 OWR43542.1 OWR43539.1 PZC75545.1 AXCN02000753 ODYU01004659 SOQ44781.1 GFDL01006655 JAV28390.1 GEZM01013099 JAV92752.1 GEZM01013100 JAV92751.1 AXCM01000824 AXCM01000825 APCN01004535 DS232158 EDS36323.1 BABH01007549 BABH01007550 CP012523 ALC39020.1 GEDC01029280 JAS08018.1 JXUM01076722 JXUM01076723 JXUM01076724 JXUM01076725 JXUM01076726 KQ562964 KXJ74746.1 UFQT01000635 SSX25968.1 CM000157 EDW88600.1 KRJ97932.1 CH477692 EAT37204.1 EAT37205.1 KQ971307 EFA11216.1 CH480818 EDW52215.1 CH954177 EDV58320.1 KQS70361.1 CM000361 CM002910 EDX04315.1 KMY89194.1 CH902624 EDV32973.1 AE014134 AY069364 AAF52731.1 AAL39509.1 AAN10684.1 AJVK01002126 CH963857 EDW76201.1 GECU01022043 GECU01006981 JAS85663.1 JAT00726.1 CH940649 EDW63241.1 KRF80986.1 CH379062 EAL32888.2 KRT05046.1 GFDF01011046 JAV03038.1 OUUW01000010 SPP85834.1 CH479184 EDW37398.1 GFDF01011234 JAV02850.1 GAIX01000062 JAA92498.1 CH916379 EDV94060.1 PZC75539.1 GAMC01015267 GAMC01015266 JAB91288.1 KB632399 ERL94647.1 APGK01039147 KB740967 ENN76865.1 CH933807 EDW12012.2 ADMH02000487 ETN66232.1 GBXI01016065 JAC98226.1
KQ460226 KPJ16448.1 GDQN01004690 JAT86364.1 AGBW02013358 OWR43540.1 PCG75607.1 JTDY01001287 KOB74293.1 ODYU01000378 SOQ35059.1 KPJ16447.1 PCG75594.1 GDQN01000412 JAT90642.1 KZ149992 PZC75543.1 PZC75541.1 KPI95042.1 PZC75542.1 KPJ16446.1 PCG75608.1 SOQ35058.1 PCG75593.1 KPJ16449.1 KPI95044.1 PCG75597.1 OWR43542.1 OWR43539.1 PZC75545.1 AXCN02000753 ODYU01004659 SOQ44781.1 GFDL01006655 JAV28390.1 GEZM01013099 JAV92752.1 GEZM01013100 JAV92751.1 AXCM01000824 AXCM01000825 APCN01004535 DS232158 EDS36323.1 BABH01007549 BABH01007550 CP012523 ALC39020.1 GEDC01029280 JAS08018.1 JXUM01076722 JXUM01076723 JXUM01076724 JXUM01076725 JXUM01076726 KQ562964 KXJ74746.1 UFQT01000635 SSX25968.1 CM000157 EDW88600.1 KRJ97932.1 CH477692 EAT37204.1 EAT37205.1 KQ971307 EFA11216.1 CH480818 EDW52215.1 CH954177 EDV58320.1 KQS70361.1 CM000361 CM002910 EDX04315.1 KMY89194.1 CH902624 EDV32973.1 AE014134 AY069364 AAF52731.1 AAL39509.1 AAN10684.1 AJVK01002126 CH963857 EDW76201.1 GECU01022043 GECU01006981 JAS85663.1 JAT00726.1 CH940649 EDW63241.1 KRF80986.1 CH379062 EAL32888.2 KRT05046.1 GFDF01011046 JAV03038.1 OUUW01000010 SPP85834.1 CH479184 EDW37398.1 GFDF01011234 JAV02850.1 GAIX01000062 JAA92498.1 CH916379 EDV94060.1 PZC75539.1 GAMC01015267 GAMC01015266 JAB91288.1 KB632399 ERL94647.1 APGK01039147 KB740967 ENN76865.1 CH933807 EDW12012.2 ADMH02000487 ETN66232.1 GBXI01016065 JAC98226.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000037510
+ More
UP000075886 UP000069272 UP000076408 UP000075880 UP000075883 UP000075903 UP000075840 UP000075882 UP000192223 UP000075900 UP000002320 UP000092553 UP000069940 UP000249989 UP000002282 UP000008820 UP000007266 UP000001292 UP000008711 UP000000304 UP000076407 UP000007801 UP000000803 UP000092462 UP000192221 UP000007798 UP000008792 UP000001819 UP000268350 UP000008744 UP000095301 UP000001070 UP000075920 UP000030742 UP000019118 UP000009192 UP000000673 UP000075885
UP000075886 UP000069272 UP000076408 UP000075880 UP000075883 UP000075903 UP000075840 UP000075882 UP000192223 UP000075900 UP000002320 UP000092553 UP000069940 UP000249989 UP000002282 UP000008820 UP000007266 UP000001292 UP000008711 UP000000304 UP000076407 UP000007801 UP000000803 UP000092462 UP000192221 UP000007798 UP000008792 UP000001819 UP000268350 UP000008744 UP000095301 UP000001070 UP000075920 UP000030742 UP000019118 UP000009192 UP000000673 UP000075885
Pfam
PF04750 Far-17a_AIG1
SUPFAM
SSF103473
SSF103473
ProteinModelPortal
H9J215
A0A194PPZ8
A0A2A4JU86
A0A0N0PDH4
A0A1E1WH93
A0A212EPX0
+ More
A0A2A4JUA0 A0A0L7LFH1 A0A2H1V2M7 A0A0N1PGW6 A0A2A4JVE8 A0A1E1WUI9 A0A2W1BUW2 A0A2W1BMM1 A0A194PP05 A0A2W1BKD6 A0A0N1IPM0 A0A2A4JV90 A0A2H1V2I5 A0A2A4JVT0 A0A0N0PDS5 A0A194PQM7 A0A2A4JU90 A0A212EPW8 A0A212EPY3 A0A2W1BPQ3 A0A182QXJ3 A0A182F236 A0A2H1VVB3 A0A182Y5M5 A0A182IPP2 A0A1Q3FL99 A0A1Y1N4E4 A0A1Y1N4B9 A0A182MSD6 A0A182UZG0 A0A1Y9GLK9 A0A182L5X9 A0A1W4WQF2 A0A182RRG8 B0WX62 H9J216 A0A0M4EQI5 A0A1B6C3D6 A0A182GNM4 A0A336MIU4 B4NXK9 Q16S04 D6W7E7 B4HYT5 B3N7V9 B4Q7C1 A0A182XCJ4 B3MUQ8 Q9VLG1 A0A1B0CZ86 A0A1W4VZN2 B4MUK6 A0A1B6JNH2 B4LUZ2 Q29JS3 A0A0R3P206 A0A1L8D9A3 A0A3B0JUR8 B4GJ84 A0A1L8D935 S4PPR9 A0A1I8NB47 B4JZD8 A0A182WHK1 A0A2W1BKH1 W8B2M2 U4UNE7 N6U5I2 B4KFB2 W5JPB6 A0A0A1WH74 A0A182PB14
A0A2A4JUA0 A0A0L7LFH1 A0A2H1V2M7 A0A0N1PGW6 A0A2A4JVE8 A0A1E1WUI9 A0A2W1BUW2 A0A2W1BMM1 A0A194PP05 A0A2W1BKD6 A0A0N1IPM0 A0A2A4JV90 A0A2H1V2I5 A0A2A4JVT0 A0A0N0PDS5 A0A194PQM7 A0A2A4JU90 A0A212EPW8 A0A212EPY3 A0A2W1BPQ3 A0A182QXJ3 A0A182F236 A0A2H1VVB3 A0A182Y5M5 A0A182IPP2 A0A1Q3FL99 A0A1Y1N4E4 A0A1Y1N4B9 A0A182MSD6 A0A182UZG0 A0A1Y9GLK9 A0A182L5X9 A0A1W4WQF2 A0A182RRG8 B0WX62 H9J216 A0A0M4EQI5 A0A1B6C3D6 A0A182GNM4 A0A336MIU4 B4NXK9 Q16S04 D6W7E7 B4HYT5 B3N7V9 B4Q7C1 A0A182XCJ4 B3MUQ8 Q9VLG1 A0A1B0CZ86 A0A1W4VZN2 B4MUK6 A0A1B6JNH2 B4LUZ2 Q29JS3 A0A0R3P206 A0A1L8D9A3 A0A3B0JUR8 B4GJ84 A0A1L8D935 S4PPR9 A0A1I8NB47 B4JZD8 A0A182WHK1 A0A2W1BKH1 W8B2M2 U4UNE7 N6U5I2 B4KFB2 W5JPB6 A0A0A1WH74 A0A182PB14
Ontologies
GO
PANTHER
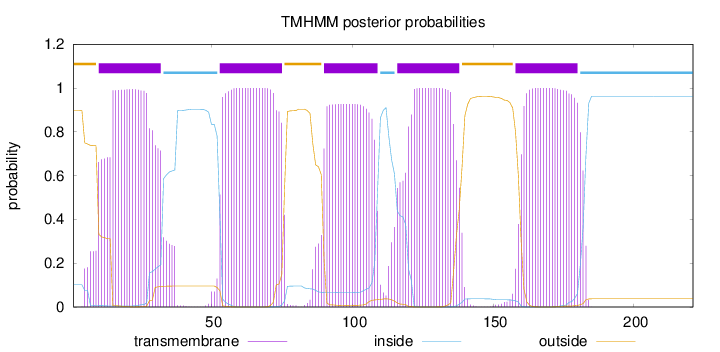
Topology
Length:
221
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
108.49503
Exp number, first 60 AAs:
30.38847
Total prob of N-in:
0.10123
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 52
TMhelix
53 - 75
outside
76 - 89
TMhelix
90 - 109
inside
110 - 115
TMhelix
116 - 138
outside
139 - 157
TMhelix
158 - 180
inside
181 - 221
Population Genetic Test Statistics
Pi
281.543324
Theta
205.721417
Tajima's D
1.153044
CLR
0.005205
CSRT
0.694315284235788
Interpretation
Uncertain
