Gene
KWMTBOMO02923
Pre Gene Modal
BGIBMGA003552
Annotation
PREDICTED:_protein_rolling_stone-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.91
Sequence
CDS
ATGACTTTCGGGTACTGGACAATATATTTAACTCATTGGGGAATTATTCTAATTACAATCACGTCGGGAGCGGCCCTCGCAGTTTCTTTGGTATCATTAAAAACTGGATGTATAGAAGCATCATTTGTACTACCGTGGTACGTGAAAGCGTACTGGGTGTGCTTCAATATCACAGTTCCTATTGCGTTTTTTATCACGATATTCTACTGGAGCTTACTGACCAAAACAGAAGATGAATATGCTATGGATCCGATTCTAGACGTGTTCATCCATGCTGTTAATGCGATCCTGATGTTAGTTCTACTAATGTTCTCTCGTCATCCTGTACGATGTCTACATTTCCACCATCCGCTATTGCTCGGAGTCGTATATCTCATCTTCAGCATTGTTTATTACTACGCTGGCGGAACTAATCCCTTTGGTGAGCCGTATGTGTACCCGATGCTGGACTGGTCGAATCCTGGGGTGGCTGCCATTACAACTGTGGGCTCGGCAATATTTATCATCATAGTACACATCATAATTGTTCTCTTGTCAGTTGCAAGGGACAGCCTTACAAGACGTTTCACGAAGTACCCACACTCATTGGACATATCTAATGACCTATACCATTAA
Protein
MTFGYWTIYLTHWGIILITITSGAALAVSLVSLKTGCIEASFVLPWYVKAYWVCFNITVPIAFFITIFYWSLLTKTEDEYAMDPILDVFIHAVNAILMLVLLMFSRHPVRCLHFHHPLLLGVVYLIFSIVYYYAGGTNPFGEPYVYPMLDWSNPGVAAITTVGSAIFIIIVHIIIVLLSVARDSLTRRFTKYPHSLDISNDLYH
Summary
Uniprot
A0A1E1WUI9
A0A2A4JVE8
A0A2W1BUW2
A0A0N1PGW6
A0A2A4JUA0
A0A2H1V2M7
+ More
A0A2H1V2I5 A0A2A4JV90 A0A0L7LFH1 A0A194PP05 A0A2W1BMM1 A0A2A4JU86 A0A212EPX0 A0A2A4JVT0 A0A194PPZ8 A0A2W1BKD6 A0A0N1IPM0 A0A0N0PDH4 A0A1E1WH93 A0A212EQ06 A0A194PNN2 A0A212EPW8 A0A0N0PDS5 A0A212EPY3 A0A194PQM7 H9J216 A0A1W4WQF2 A0A2W1BPQ3 A0A182F236 A0A182MSD6 A0A2A4JU90 A0A0A1WWB7 A0A182QXJ3 W8BXI1 A0A0A1X9A3 A0A182Y5M5 A0A182UZG0 A0A1Y9GLK9 A0A182L5X9 A0A1L8D9A3 A0A182IPP2 A0A182RRG8 A0A1L8D935 A0A2H1VVB3 A0A1Q3FL99 A0A1I8P1F6 A0A1B0CZ86 B0WX62 A0A1Y1N4E4 A0A1Y1N4B9 D6W7E7 Q16S04 A0A1I8N097 A0A1B6C3D6 K1Q5N2 A0A182GNM4 A0A2H1V2K1 A0A2W1BKH1 A0A034W4U6 A0A0T6AYG0 A0A1I8PR73 A0A0L0CQ92 A0A2A4JV12 A0A0L0CMJ3 A0A0K8TRE5 A0A212F2Q7 A0A1W4VZN2 B3MUQ8 B4KFB4 B5DKA8 D6QYP2 B4GJ86 D6QYP8 B4NXK9 A0A336MIU4 A7SMX4 A0A1B0FKK2 A0A0A1WSZ7 A0A1B6JNH2 C3YTA7 V4AHJ2 B4JZD6 A0A1I8MB35 A0A1A9XWS7 A0A1B0BP37 D6QYR0 A0A3B0JUP9 A0A1I8PM89 A0A1A9ZHP5 B3N7V9
A0A2H1V2I5 A0A2A4JV90 A0A0L7LFH1 A0A194PP05 A0A2W1BMM1 A0A2A4JU86 A0A212EPX0 A0A2A4JVT0 A0A194PPZ8 A0A2W1BKD6 A0A0N1IPM0 A0A0N0PDH4 A0A1E1WH93 A0A212EQ06 A0A194PNN2 A0A212EPW8 A0A0N0PDS5 A0A212EPY3 A0A194PQM7 H9J216 A0A1W4WQF2 A0A2W1BPQ3 A0A182F236 A0A182MSD6 A0A2A4JU90 A0A0A1WWB7 A0A182QXJ3 W8BXI1 A0A0A1X9A3 A0A182Y5M5 A0A182UZG0 A0A1Y9GLK9 A0A182L5X9 A0A1L8D9A3 A0A182IPP2 A0A182RRG8 A0A1L8D935 A0A2H1VVB3 A0A1Q3FL99 A0A1I8P1F6 A0A1B0CZ86 B0WX62 A0A1Y1N4E4 A0A1Y1N4B9 D6W7E7 Q16S04 A0A1I8N097 A0A1B6C3D6 K1Q5N2 A0A182GNM4 A0A2H1V2K1 A0A2W1BKH1 A0A034W4U6 A0A0T6AYG0 A0A1I8PR73 A0A0L0CQ92 A0A2A4JV12 A0A0L0CMJ3 A0A0K8TRE5 A0A212F2Q7 A0A1W4VZN2 B3MUQ8 B4KFB4 B5DKA8 D6QYP2 B4GJ86 D6QYP8 B4NXK9 A0A336MIU4 A7SMX4 A0A1B0FKK2 A0A0A1WSZ7 A0A1B6JNH2 C3YTA7 V4AHJ2 B4JZD6 A0A1I8MB35 A0A1A9XWS7 A0A1B0BP37 D6QYR0 A0A3B0JUP9 A0A1I8PM89 A0A1A9ZHP5 B3N7V9
Pubmed
EMBL
GDQN01000412
JAT90642.1
NWSH01000568
PCG75594.1
KZ149992
PZC75543.1
+ More
KQ460226 KPJ16447.1 PCG75607.1 ODYU01000378 SOQ35059.1 SOQ35058.1 PCG75608.1 JTDY01001287 KOB74293.1 KQ459597 KPI95042.1 PZC75541.1 PCG75595.1 AGBW02013358 OWR43540.1 PCG75593.1 KPI95043.1 PZC75542.1 KPJ16446.1 KPJ16448.1 GDQN01004690 JAT86364.1 OWR43541.1 KPI95041.1 OWR43542.1 KPJ16449.1 OWR43539.1 KPI95044.1 BABH01007549 BABH01007550 PZC75545.1 AXCM01000824 AXCM01000825 PCG75597.1 GBXI01010963 JAD03329.1 AXCN02000753 GAMC01000485 JAC06071.1 GBXI01007029 JAD07263.1 APCN01004535 GFDF01011046 JAV03038.1 GFDF01011234 JAV02850.1 ODYU01004659 SOQ44781.1 GFDL01006655 JAV28390.1 AJVK01002126 DS232158 EDS36323.1 GEZM01013099 JAV92752.1 GEZM01013100 JAV92751.1 KQ971307 EFA11216.1 CH477692 EAT37204.1 EAT37205.1 GEDC01029280 JAS08018.1 JH817548 EKC29218.1 JXUM01076722 JXUM01076723 JXUM01076724 JXUM01076725 JXUM01076726 KQ562964 KXJ74746.1 SOQ35061.1 PZC75539.1 GAKP01008336 JAC50616.1 LJIG01022567 KRT79885.1 JRES01000175 KNC33609.1 PCG75606.1 KNC33573.1 GDAI01001108 JAI16495.1 AGBW02010703 OWR48003.1 CH902624 EDV32973.1 CH933807 EDW12014.1 CH379062 EDY70734.1 HM017711 HM017712 HM017713 HM017714 HM017715 HM017716 HM017718 HM017719 HM017720 HM017721 ADG65062.1 ADG65063.1 ADG65064.1 ADG65065.1 ADG65066.1 ADG65067.1 ADG65069.1 ADG65070.1 ADG65071.1 ADG65072.1 CH479184 EDW37400.1 HM017717 ADG65068.1 CM000157 EDW88600.1 KRJ97932.1 UFQT01000635 SSX25968.1 DS469714 EDO34937.1 CCAG010008391 GBXI01012325 JAD01967.1 GECU01022043 GECU01006981 JAS85663.1 JAT00726.1 GG666551 EEN56474.1 KB199981 ESP03529.1 CH916379 EDV94058.1 JXJN01017803 HM017729 ADG65080.1 OUUW01000010 SPP85837.1 CH954177 EDV58320.1 KQS70361.1
KQ460226 KPJ16447.1 PCG75607.1 ODYU01000378 SOQ35059.1 SOQ35058.1 PCG75608.1 JTDY01001287 KOB74293.1 KQ459597 KPI95042.1 PZC75541.1 PCG75595.1 AGBW02013358 OWR43540.1 PCG75593.1 KPI95043.1 PZC75542.1 KPJ16446.1 KPJ16448.1 GDQN01004690 JAT86364.1 OWR43541.1 KPI95041.1 OWR43542.1 KPJ16449.1 OWR43539.1 KPI95044.1 BABH01007549 BABH01007550 PZC75545.1 AXCM01000824 AXCM01000825 PCG75597.1 GBXI01010963 JAD03329.1 AXCN02000753 GAMC01000485 JAC06071.1 GBXI01007029 JAD07263.1 APCN01004535 GFDF01011046 JAV03038.1 GFDF01011234 JAV02850.1 ODYU01004659 SOQ44781.1 GFDL01006655 JAV28390.1 AJVK01002126 DS232158 EDS36323.1 GEZM01013099 JAV92752.1 GEZM01013100 JAV92751.1 KQ971307 EFA11216.1 CH477692 EAT37204.1 EAT37205.1 GEDC01029280 JAS08018.1 JH817548 EKC29218.1 JXUM01076722 JXUM01076723 JXUM01076724 JXUM01076725 JXUM01076726 KQ562964 KXJ74746.1 SOQ35061.1 PZC75539.1 GAKP01008336 JAC50616.1 LJIG01022567 KRT79885.1 JRES01000175 KNC33609.1 PCG75606.1 KNC33573.1 GDAI01001108 JAI16495.1 AGBW02010703 OWR48003.1 CH902624 EDV32973.1 CH933807 EDW12014.1 CH379062 EDY70734.1 HM017711 HM017712 HM017713 HM017714 HM017715 HM017716 HM017718 HM017719 HM017720 HM017721 ADG65062.1 ADG65063.1 ADG65064.1 ADG65065.1 ADG65066.1 ADG65067.1 ADG65069.1 ADG65070.1 ADG65071.1 ADG65072.1 CH479184 EDW37400.1 HM017717 ADG65068.1 CM000157 EDW88600.1 KRJ97932.1 UFQT01000635 SSX25968.1 DS469714 EDO34937.1 CCAG010008391 GBXI01012325 JAD01967.1 GECU01022043 GECU01006981 JAS85663.1 JAT00726.1 GG666551 EEN56474.1 KB199981 ESP03529.1 CH916379 EDV94058.1 JXJN01017803 HM017729 ADG65080.1 OUUW01000010 SPP85837.1 CH954177 EDV58320.1 KQS70361.1
Proteomes
UP000218220
UP000053240
UP000037510
UP000053268
UP000007151
UP000005204
+ More
UP000192223 UP000069272 UP000075883 UP000075886 UP000076408 UP000075903 UP000075840 UP000075882 UP000075880 UP000075900 UP000095300 UP000092462 UP000002320 UP000007266 UP000008820 UP000095301 UP000005408 UP000069940 UP000249989 UP000037069 UP000192221 UP000007801 UP000009192 UP000001819 UP000008744 UP000002282 UP000001593 UP000092444 UP000001554 UP000030746 UP000001070 UP000092443 UP000092460 UP000268350 UP000092445 UP000008711
UP000192223 UP000069272 UP000075883 UP000075886 UP000076408 UP000075903 UP000075840 UP000075882 UP000075880 UP000075900 UP000095300 UP000092462 UP000002320 UP000007266 UP000008820 UP000095301 UP000005408 UP000069940 UP000249989 UP000037069 UP000192221 UP000007801 UP000009192 UP000001819 UP000008744 UP000002282 UP000001593 UP000092444 UP000001554 UP000030746 UP000001070 UP000092443 UP000092460 UP000268350 UP000092445 UP000008711
PRIDE
Pfam
PF04750 Far-17a_AIG1
SUPFAM
SSF103473
SSF103473
ProteinModelPortal
A0A1E1WUI9
A0A2A4JVE8
A0A2W1BUW2
A0A0N1PGW6
A0A2A4JUA0
A0A2H1V2M7
+ More
A0A2H1V2I5 A0A2A4JV90 A0A0L7LFH1 A0A194PP05 A0A2W1BMM1 A0A2A4JU86 A0A212EPX0 A0A2A4JVT0 A0A194PPZ8 A0A2W1BKD6 A0A0N1IPM0 A0A0N0PDH4 A0A1E1WH93 A0A212EQ06 A0A194PNN2 A0A212EPW8 A0A0N0PDS5 A0A212EPY3 A0A194PQM7 H9J216 A0A1W4WQF2 A0A2W1BPQ3 A0A182F236 A0A182MSD6 A0A2A4JU90 A0A0A1WWB7 A0A182QXJ3 W8BXI1 A0A0A1X9A3 A0A182Y5M5 A0A182UZG0 A0A1Y9GLK9 A0A182L5X9 A0A1L8D9A3 A0A182IPP2 A0A182RRG8 A0A1L8D935 A0A2H1VVB3 A0A1Q3FL99 A0A1I8P1F6 A0A1B0CZ86 B0WX62 A0A1Y1N4E4 A0A1Y1N4B9 D6W7E7 Q16S04 A0A1I8N097 A0A1B6C3D6 K1Q5N2 A0A182GNM4 A0A2H1V2K1 A0A2W1BKH1 A0A034W4U6 A0A0T6AYG0 A0A1I8PR73 A0A0L0CQ92 A0A2A4JV12 A0A0L0CMJ3 A0A0K8TRE5 A0A212F2Q7 A0A1W4VZN2 B3MUQ8 B4KFB4 B5DKA8 D6QYP2 B4GJ86 D6QYP8 B4NXK9 A0A336MIU4 A7SMX4 A0A1B0FKK2 A0A0A1WSZ7 A0A1B6JNH2 C3YTA7 V4AHJ2 B4JZD6 A0A1I8MB35 A0A1A9XWS7 A0A1B0BP37 D6QYR0 A0A3B0JUP9 A0A1I8PM89 A0A1A9ZHP5 B3N7V9
A0A2H1V2I5 A0A2A4JV90 A0A0L7LFH1 A0A194PP05 A0A2W1BMM1 A0A2A4JU86 A0A212EPX0 A0A2A4JVT0 A0A194PPZ8 A0A2W1BKD6 A0A0N1IPM0 A0A0N0PDH4 A0A1E1WH93 A0A212EQ06 A0A194PNN2 A0A212EPW8 A0A0N0PDS5 A0A212EPY3 A0A194PQM7 H9J216 A0A1W4WQF2 A0A2W1BPQ3 A0A182F236 A0A182MSD6 A0A2A4JU90 A0A0A1WWB7 A0A182QXJ3 W8BXI1 A0A0A1X9A3 A0A182Y5M5 A0A182UZG0 A0A1Y9GLK9 A0A182L5X9 A0A1L8D9A3 A0A182IPP2 A0A182RRG8 A0A1L8D935 A0A2H1VVB3 A0A1Q3FL99 A0A1I8P1F6 A0A1B0CZ86 B0WX62 A0A1Y1N4E4 A0A1Y1N4B9 D6W7E7 Q16S04 A0A1I8N097 A0A1B6C3D6 K1Q5N2 A0A182GNM4 A0A2H1V2K1 A0A2W1BKH1 A0A034W4U6 A0A0T6AYG0 A0A1I8PR73 A0A0L0CQ92 A0A2A4JV12 A0A0L0CMJ3 A0A0K8TRE5 A0A212F2Q7 A0A1W4VZN2 B3MUQ8 B4KFB4 B5DKA8 D6QYP2 B4GJ86 D6QYP8 B4NXK9 A0A336MIU4 A7SMX4 A0A1B0FKK2 A0A0A1WSZ7 A0A1B6JNH2 C3YTA7 V4AHJ2 B4JZD6 A0A1I8MB35 A0A1A9XWS7 A0A1B0BP37 D6QYR0 A0A3B0JUP9 A0A1I8PM89 A0A1A9ZHP5 B3N7V9
Ontologies
PANTHER
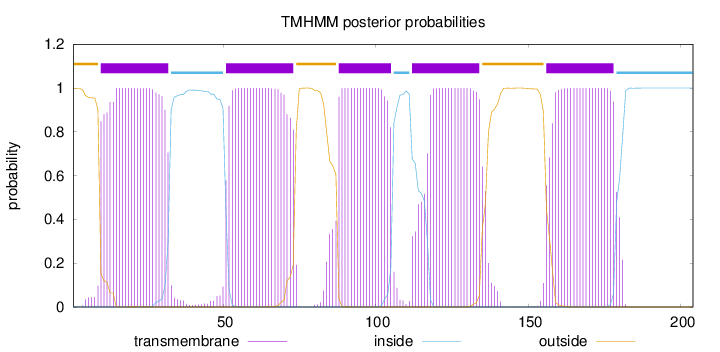
Topology
Length:
204
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
109.10826
Exp number, first 60 AAs:
32.32696
Total prob of N-in:
0.00170
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 50
TMhelix
51 - 73
outside
74 - 87
TMhelix
88 - 105
inside
106 - 111
TMhelix
112 - 134
outside
135 - 155
TMhelix
156 - 178
inside
179 - 204
Population Genetic Test Statistics
Pi
231.242171
Theta
160.932183
Tajima's D
1.504633
CLR
0.000893
CSRT
0.787960601969901
Interpretation
Uncertain
