Gene
KWMTBOMO02922
Annotation
PREDICTED:_protein_rolling_stone-like_isoform_X3_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.973
Sequence
CDS
ATGATCAAATACACATGGAAACACGGATTGTACTGCATCTCGGCATTTCTGGGATCGGAAGATGAATCCGTCGAGATCTTTACGAAATCCAGCTTGGGAAGCATTTCCCCTTTGTTATGGAGGATACCAATATTCTTGTGGGCAACCATCTCACTTTTCATGTCGATGAATTATTTCTGGGGGCCCACGAAGCTATTCTTTGTGTATATGACACATTGGGGTCTGTTATTCATCGTCCTGGAATCTATGTTCGGTATCATCGTAGTACTAAGGAAACGCGGAAGACCAGACGCTACATTTGGCTTGCCTTGGTATGTAAAAACGTACTGGGTCCTCTACAATATTTCAATCCCGGTCGCCTTTCTCATTACTGTGTTCTATTGGGGAATATTAAAATCATCATTGGACACAGTGAAGTTCTCACCGAATCCTGTGCTGGACGTAATGATTCACGGCGTGAACTCCGTTGTAATGTTCGTTGAGCTGATGTTCTCGGCGCATCCCAGTAGACTGCTACACATCATGCAACCGCTGTACTTCGCCGGAGCCTACATGCTATTCTCTGTCACTTATTATTTCGCTGGTGGATTAGATCCCTGGGGGAACAAATTCATCTATCCAGTGATAGACTGGTCAAAGCCAGATCAAACGATGGTTGTAATTACCCTCACGGCTATATTCCTTGCCTTAATGCATCTCATCACCGTGGCAATGGCGGCTGCTCGGGACTCCATTACGAAACAATGTAAACCAGATCCAACAGGAGTTTACAATGATGGATTCGAACCCTAA
Protein
MIKYTWKHGLYCISAFLGSEDESVEIFTKSSLGSISPLLWRIPIFLWATISLFMSMNYFWGPTKLFFVYMTHWGLLFIVLESMFGIIVVLRKRGRPDATFGLPWYVKTYWVLYNISIPVAFLITVFYWGILKSSLDTVKFSPNPVLDVMIHGVNSVVMFVELMFSAHPSRLLHIMQPLYFAGAYMLFSVTYYFAGGLDPWGNKFIYPVIDWSKPDQTMVVITLTAIFLALMHLITVAMAAARDSITKQCKPDPTGVYNDGFEP
Summary
Uniprot
A0A2W1BMM1
A0A0N1IPM0
A0A2A4JUA0
A0A2H1V2M7
A0A0L7LFH1
A0A194PNN2
+ More
A0A212EPX0 H9J215 A0A194PPZ8 A0A1E1WUI9 A0A2A4JVE8 A0A2A4JU86 A0A1E1WH93 A0A0N0PDH4 A0A2W1BKD6 A0A2A4JV90 A0A2A4JVT0 A0A0N1PGW6 A0A194PP05 A0A212EPY3 A0A182RRG8 A0A182Y5M5 A0A182MSD6 A0A1Y9GLK9 A0A182L5X9 A0A182UZG0 A0A182F236 A0A194PQM7 A0A182IPP2 A0A182QXJ3 A0A2A4JU90 A0A0N0PDS5 A0A2W1BPQ3 A0A336MIU4 A0A0L0CMJ3 A0A1I8P1F6 D6W7E7 A0A1B6C3D6 A0A212EQ06 H9J216 N6U5I2 U4UNE7 A0A1W4VZN2 B4JZD6 B4MUK6 B4Q7C1 B4HYT5 A0A1W4WQF2 Q9VLG1 B3N7V9 B4NXK9 A0A2H1V2K1 A0A1S3H9V7 A0A0K8TRE5 A0A2W1BKH1 B4JZD8 A0A1B6JNH2 A0A3B0JUR8 A0A0C9QXI8 A0A0R3P206 A0A0C9PVD2 Q29JS3 A0A1Y1N4B9 B4LUZ2 A0A1Y1N4E4 A0A026WND4 A0A212F2Q7 A0A1A9UJM3 A0A1A9YBQ8 A0A0M4EQI5 A0A3L8DHS8 A0A1B0AQ44 A0A1I8N097 B4LUZ4 B4JAV0 A0A0M4EBX1 B4GJ84 A0A1I8NDS0
A0A212EPX0 H9J215 A0A194PPZ8 A0A1E1WUI9 A0A2A4JVE8 A0A2A4JU86 A0A1E1WH93 A0A0N0PDH4 A0A2W1BKD6 A0A2A4JV90 A0A2A4JVT0 A0A0N1PGW6 A0A194PP05 A0A212EPY3 A0A182RRG8 A0A182Y5M5 A0A182MSD6 A0A1Y9GLK9 A0A182L5X9 A0A182UZG0 A0A182F236 A0A194PQM7 A0A182IPP2 A0A182QXJ3 A0A2A4JU90 A0A0N0PDS5 A0A2W1BPQ3 A0A336MIU4 A0A0L0CMJ3 A0A1I8P1F6 D6W7E7 A0A1B6C3D6 A0A212EQ06 H9J216 N6U5I2 U4UNE7 A0A1W4VZN2 B4JZD6 B4MUK6 B4Q7C1 B4HYT5 A0A1W4WQF2 Q9VLG1 B3N7V9 B4NXK9 A0A2H1V2K1 A0A1S3H9V7 A0A0K8TRE5 A0A2W1BKH1 B4JZD8 A0A1B6JNH2 A0A3B0JUR8 A0A0C9QXI8 A0A0R3P206 A0A0C9PVD2 Q29JS3 A0A1Y1N4B9 B4LUZ2 A0A1Y1N4E4 A0A026WND4 A0A212F2Q7 A0A1A9UJM3 A0A1A9YBQ8 A0A0M4EQI5 A0A3L8DHS8 A0A1B0AQ44 A0A1I8N097 B4LUZ4 B4JAV0 A0A0M4EBX1 B4GJ84 A0A1I8NDS0
Pubmed
EMBL
KZ149992
PZC75541.1
KQ460226
KPJ16446.1
NWSH01000568
PCG75607.1
+ More
ODYU01000378 SOQ35059.1 JTDY01001287 KOB74293.1 KQ459597 KPI95041.1 AGBW02013358 OWR43540.1 BABH01007552 BABH01007553 KPI95043.1 GDQN01000412 JAT90642.1 PCG75594.1 PCG75595.1 GDQN01004690 JAT86364.1 KPJ16448.1 PZC75542.1 PCG75608.1 PCG75593.1 KPJ16447.1 KPI95042.1 OWR43539.1 AXCM01000824 AXCM01000825 APCN01004535 KPI95044.1 AXCN02000753 PCG75597.1 KPJ16449.1 PZC75545.1 UFQT01000635 SSX25968.1 JRES01000175 KNC33573.1 KQ971307 EFA11216.1 GEDC01029280 JAS08018.1 OWR43541.1 BABH01007549 BABH01007550 APGK01039147 KB740967 ENN76865.1 KB632399 ERL94647.1 CH916379 EDV94058.1 CH963857 EDW76201.1 CM000361 CM002910 EDX04315.1 KMY89194.1 CH480818 EDW52215.1 AE014134 AY069364 AAF52731.1 AAL39509.1 AAN10684.1 CH954177 EDV58320.1 KQS70361.1 CM000157 EDW88600.1 KRJ97932.1 SOQ35061.1 GDAI01001108 JAI16495.1 PZC75539.1 EDV94060.1 GECU01022043 GECU01006981 JAS85663.1 JAT00726.1 OUUW01000010 SPP85834.1 GBYB01005387 JAG75154.1 CH379062 KRT05046.1 GBYB01005388 JAG75155.1 EAL32888.2 GEZM01013100 JAV92751.1 CH940649 EDW63241.1 KRF80986.1 GEZM01013099 JAV92752.1 KK107152 EZA57146.1 AGBW02010703 OWR48003.1 CP012523 ALC39020.1 QOIP01000007 RLU20005.1 JXJN01001669 EDW63243.1 CH916368 EDW02820.1 ALC39022.1 CH479184 EDW37398.1
ODYU01000378 SOQ35059.1 JTDY01001287 KOB74293.1 KQ459597 KPI95041.1 AGBW02013358 OWR43540.1 BABH01007552 BABH01007553 KPI95043.1 GDQN01000412 JAT90642.1 PCG75594.1 PCG75595.1 GDQN01004690 JAT86364.1 KPJ16448.1 PZC75542.1 PCG75608.1 PCG75593.1 KPJ16447.1 KPI95042.1 OWR43539.1 AXCM01000824 AXCM01000825 APCN01004535 KPI95044.1 AXCN02000753 PCG75597.1 KPJ16449.1 PZC75545.1 UFQT01000635 SSX25968.1 JRES01000175 KNC33573.1 KQ971307 EFA11216.1 GEDC01029280 JAS08018.1 OWR43541.1 BABH01007549 BABH01007550 APGK01039147 KB740967 ENN76865.1 KB632399 ERL94647.1 CH916379 EDV94058.1 CH963857 EDW76201.1 CM000361 CM002910 EDX04315.1 KMY89194.1 CH480818 EDW52215.1 AE014134 AY069364 AAF52731.1 AAL39509.1 AAN10684.1 CH954177 EDV58320.1 KQS70361.1 CM000157 EDW88600.1 KRJ97932.1 SOQ35061.1 GDAI01001108 JAI16495.1 PZC75539.1 EDV94060.1 GECU01022043 GECU01006981 JAS85663.1 JAT00726.1 OUUW01000010 SPP85834.1 GBYB01005387 JAG75154.1 CH379062 KRT05046.1 GBYB01005388 JAG75155.1 EAL32888.2 GEZM01013100 JAV92751.1 CH940649 EDW63241.1 KRF80986.1 GEZM01013099 JAV92752.1 KK107152 EZA57146.1 AGBW02010703 OWR48003.1 CP012523 ALC39020.1 QOIP01000007 RLU20005.1 JXJN01001669 EDW63243.1 CH916368 EDW02820.1 ALC39022.1 CH479184 EDW37398.1
Proteomes
UP000053240
UP000218220
UP000037510
UP000053268
UP000007151
UP000005204
+ More
UP000075900 UP000076408 UP000075883 UP000075840 UP000075882 UP000075903 UP000069272 UP000075880 UP000075886 UP000037069 UP000095300 UP000007266 UP000019118 UP000030742 UP000192221 UP000001070 UP000007798 UP000000304 UP000001292 UP000192223 UP000000803 UP000008711 UP000002282 UP000085678 UP000268350 UP000001819 UP000008792 UP000053097 UP000078200 UP000092443 UP000092553 UP000279307 UP000092460 UP000095301 UP000008744
UP000075900 UP000076408 UP000075883 UP000075840 UP000075882 UP000075903 UP000069272 UP000075880 UP000075886 UP000037069 UP000095300 UP000007266 UP000019118 UP000030742 UP000192221 UP000001070 UP000007798 UP000000304 UP000001292 UP000192223 UP000000803 UP000008711 UP000002282 UP000085678 UP000268350 UP000001819 UP000008792 UP000053097 UP000078200 UP000092443 UP000092553 UP000279307 UP000092460 UP000095301 UP000008744
Pfam
PF04750 Far-17a_AIG1
SUPFAM
SSF103473
SSF103473
ProteinModelPortal
A0A2W1BMM1
A0A0N1IPM0
A0A2A4JUA0
A0A2H1V2M7
A0A0L7LFH1
A0A194PNN2
+ More
A0A212EPX0 H9J215 A0A194PPZ8 A0A1E1WUI9 A0A2A4JVE8 A0A2A4JU86 A0A1E1WH93 A0A0N0PDH4 A0A2W1BKD6 A0A2A4JV90 A0A2A4JVT0 A0A0N1PGW6 A0A194PP05 A0A212EPY3 A0A182RRG8 A0A182Y5M5 A0A182MSD6 A0A1Y9GLK9 A0A182L5X9 A0A182UZG0 A0A182F236 A0A194PQM7 A0A182IPP2 A0A182QXJ3 A0A2A4JU90 A0A0N0PDS5 A0A2W1BPQ3 A0A336MIU4 A0A0L0CMJ3 A0A1I8P1F6 D6W7E7 A0A1B6C3D6 A0A212EQ06 H9J216 N6U5I2 U4UNE7 A0A1W4VZN2 B4JZD6 B4MUK6 B4Q7C1 B4HYT5 A0A1W4WQF2 Q9VLG1 B3N7V9 B4NXK9 A0A2H1V2K1 A0A1S3H9V7 A0A0K8TRE5 A0A2W1BKH1 B4JZD8 A0A1B6JNH2 A0A3B0JUR8 A0A0C9QXI8 A0A0R3P206 A0A0C9PVD2 Q29JS3 A0A1Y1N4B9 B4LUZ2 A0A1Y1N4E4 A0A026WND4 A0A212F2Q7 A0A1A9UJM3 A0A1A9YBQ8 A0A0M4EQI5 A0A3L8DHS8 A0A1B0AQ44 A0A1I8N097 B4LUZ4 B4JAV0 A0A0M4EBX1 B4GJ84 A0A1I8NDS0
A0A212EPX0 H9J215 A0A194PPZ8 A0A1E1WUI9 A0A2A4JVE8 A0A2A4JU86 A0A1E1WH93 A0A0N0PDH4 A0A2W1BKD6 A0A2A4JV90 A0A2A4JVT0 A0A0N1PGW6 A0A194PP05 A0A212EPY3 A0A182RRG8 A0A182Y5M5 A0A182MSD6 A0A1Y9GLK9 A0A182L5X9 A0A182UZG0 A0A182F236 A0A194PQM7 A0A182IPP2 A0A182QXJ3 A0A2A4JU90 A0A0N0PDS5 A0A2W1BPQ3 A0A336MIU4 A0A0L0CMJ3 A0A1I8P1F6 D6W7E7 A0A1B6C3D6 A0A212EQ06 H9J216 N6U5I2 U4UNE7 A0A1W4VZN2 B4JZD6 B4MUK6 B4Q7C1 B4HYT5 A0A1W4WQF2 Q9VLG1 B3N7V9 B4NXK9 A0A2H1V2K1 A0A1S3H9V7 A0A0K8TRE5 A0A2W1BKH1 B4JZD8 A0A1B6JNH2 A0A3B0JUR8 A0A0C9QXI8 A0A0R3P206 A0A0C9PVD2 Q29JS3 A0A1Y1N4B9 B4LUZ2 A0A1Y1N4E4 A0A026WND4 A0A212F2Q7 A0A1A9UJM3 A0A1A9YBQ8 A0A0M4EQI5 A0A3L8DHS8 A0A1B0AQ44 A0A1I8N097 B4LUZ4 B4JAV0 A0A0M4EBX1 B4GJ84 A0A1I8NDS0
Ontologies
PANTHER
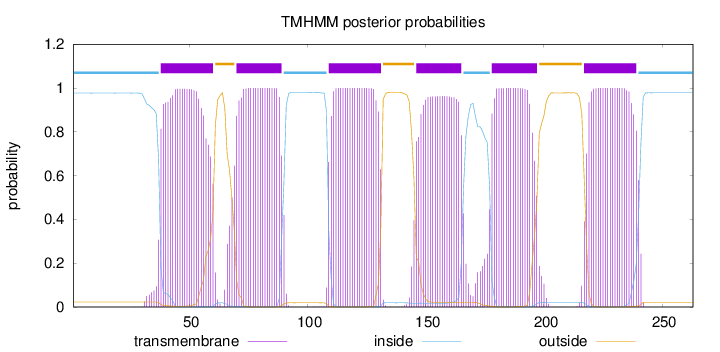
Topology
Length:
263
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
128.98299
Exp number, first 60 AAs:
21.64409
Total prob of N-in:
0.97704
POSSIBLE N-term signal
sequence
inside
1 - 37
TMhelix
38 - 60
outside
61 - 69
TMhelix
70 - 89
inside
90 - 108
TMhelix
109 - 131
outside
132 - 145
TMhelix
146 - 165
inside
166 - 177
TMhelix
178 - 197
outside
198 - 216
TMhelix
217 - 239
inside
240 - 263
Population Genetic Test Statistics
Pi
230.464915
Theta
169.222531
Tajima's D
0.737759
CLR
0.661924
CSRT
0.587270636468177
Interpretation
Uncertain
