Gene
KWMTBOMO02921
Pre Gene Modal
BGIBMGA003551
Annotation
PREDICTED:_protein_rolling_stone_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.076
Sequence
CDS
ATGCAGAAGTCTTGCAAACCAGCGATGCTTGGCCGCATGTGGCAAGCCAGTAGCCGAGCTTTTAGCTTGGATCATTGGCCTTCTCATCGCTTTGGCTGTTGCCAGTGGCAGAAGAGGCAACAACCTTCCAAATACTATATTCTGTACAGGTGGATAATATTTATTGCGGTATTGGCCATTAACATTGCGAGTTTTGCTTGTCAGCGCATCCCAGTGAAGTATCACGGACTGAAAGTGGAACTAAATTACTTTAAATGGTTCATTTACCTCACTAATTGGGGATTTATGCTGATAGCTGTTCAAGCAGCTCTCGCACTTGGCGTCGTGTACTACTATAAAAACGACAGGAATCTTAATCTAACTTACGATGAAGATGATGTTCCTGTACCGATGTCTCGTCGTAAACGTACCCCAATTTTGTGCCGCGTGTATTGGCTCACACAAAACGTCGCTACAGACCTCGCCTTCGTCATCTCTCTTGTTTACTGGGCCCTAGTGTATGACCCTGAAATCCATGAAATAAATGCAATAAATCTTCTTGTTCATGGTGGAAATTCCGCGATCATGTTGTGTGAGCTGGCTGTCACAGCACACCCCATTAGGATGGCTCATGCAATGTACGGCGCCGGAGCAGGTCTTGCTTATGGACTCTTCAGTGCATTTTACTGGGCAGTAGGGGGCACGGACAGACTTGGACAGGCTGCTATATATCCCACCCTGGATTGGAATAAACCAGCCGCTGCATTTGGCTTCGTCGCTCTGTGTGCTGTGGTGCTGGTGATGGCTCACGGCGTCGCGACCTGCCTAGCCGTAGTCCGTTTGCGTCTCGCCAAGCGCCTGTACGCAAAACGCGCGCAATCCGACAGACTCACCCTACCGACACATTGA
Protein
MQKSCKPAMLGRMWQASSRAFSLDHWPSHRFGCCQWQKRQQPSKYYILYRWIIFIAVLAINIASFACQRIPVKYHGLKVELNYFKWFIYLTNWGFMLIAVQAALALGVVYYYKNDRNLNLTYDEDDVPVPMSRRKRTPILCRVYWLTQNVATDLAFVISLVYWALVYDPEIHEINAINLLVHGGNSAIMLCELAVTAHPIRMAHAMYGAGAGLAYGLFSAFYWAVGGTDRLGQAAIYPTLDWNKPAAAFGFVALCAVVLVMAHGVATCLAVVRLRLAKRLYAKRAQSDRLTLPTH
Summary
Uniprot
H9J214
S4PPR9
A0A2A4JV12
A0A2W1BKH1
A0A194PV82
A0A0N1I933
+ More
A0A2H1V2K1 A0A212F2Q7 W8C1X8 A0A1B0A9N6 A0A1B0FBW6 A0A034VS26 A0A1A9WBX3 Q9VK03 A0A0J9R1S4 A0A0P8XHF7 A0A1I8PM89 A0A1B0AQ44 A0A1A9YBQ8 A0A1A9UJM3 A0A0Q9XDS7 M9PFU0 B4M982 B3NAY7 A0A1W4UUU2 B4P347 A0A1B6C3D6 A0A3B0KH96 A0A1I8NDS0 Q29JZ5 A0A1B6LQ50 A0A1L8D6R2 B0W430 Q17A74 A0A182QD39 A0A182NUY8 A0A182F235 A0A182RRG7 A0A336MP41 A0A182PB16 A0A182IWF9 A0A182Y5M6 A0A182WHK3 A0A1B6JNH2 A0A0Q9WQ80 A0A182L5X6 A0A182VF07 A0A182XCJ6 A0A182IDX6 A0A1B0CA98 A0A1W4WQF2 A0A2J7PJ37 A0A182TE01 A0A2J7PJ65 A0A182MUK6 A0A1J1HRB6 A0A067QSW7 A0A3B0KC43 A0A1Y1N4E4 A0A2P8ZHD3 A0A1Y1N4B9 D6W7E7 A0A0T6AYG0 A0A067RT92 A0A1B6FLK0 A0A026WND4 A0A232F2K5 A0A3L8DHS8 A0A069DS34 A0A087ZZV6 A0A0K8TRE5 A0A0A9WND3 A0A1B0CZ86 V9IHR3 A0A0A1WH74 A0A182Y5M5 A0A0J7KQI2 A0A0L0CMU8 E9IZQ9 A0A182QXJ3 A0A1L8D935 A0A034VGR5 A0A0K8V861 A0A2A4JVE8 A0A336MHR5 A0A0L7R1V2 A0A182RRG8 W8B2M2 A0A336MKK2 E2BCM1 A0A154P335 T1HR70 B0WX62 A0A0N0BDM5 A0A151X135 A0A1Q3FL99
A0A2H1V2K1 A0A212F2Q7 W8C1X8 A0A1B0A9N6 A0A1B0FBW6 A0A034VS26 A0A1A9WBX3 Q9VK03 A0A0J9R1S4 A0A0P8XHF7 A0A1I8PM89 A0A1B0AQ44 A0A1A9YBQ8 A0A1A9UJM3 A0A0Q9XDS7 M9PFU0 B4M982 B3NAY7 A0A1W4UUU2 B4P347 A0A1B6C3D6 A0A3B0KH96 A0A1I8NDS0 Q29JZ5 A0A1B6LQ50 A0A1L8D6R2 B0W430 Q17A74 A0A182QD39 A0A182NUY8 A0A182F235 A0A182RRG7 A0A336MP41 A0A182PB16 A0A182IWF9 A0A182Y5M6 A0A182WHK3 A0A1B6JNH2 A0A0Q9WQ80 A0A182L5X6 A0A182VF07 A0A182XCJ6 A0A182IDX6 A0A1B0CA98 A0A1W4WQF2 A0A2J7PJ37 A0A182TE01 A0A2J7PJ65 A0A182MUK6 A0A1J1HRB6 A0A067QSW7 A0A3B0KC43 A0A1Y1N4E4 A0A2P8ZHD3 A0A1Y1N4B9 D6W7E7 A0A0T6AYG0 A0A067RT92 A0A1B6FLK0 A0A026WND4 A0A232F2K5 A0A3L8DHS8 A0A069DS34 A0A087ZZV6 A0A0K8TRE5 A0A0A9WND3 A0A1B0CZ86 V9IHR3 A0A0A1WH74 A0A182Y5M5 A0A0J7KQI2 A0A0L0CMU8 E9IZQ9 A0A182QXJ3 A0A1L8D935 A0A034VGR5 A0A0K8V861 A0A2A4JVE8 A0A336MHR5 A0A0L7R1V2 A0A182RRG8 W8B2M2 A0A336MKK2 E2BCM1 A0A154P335 T1HR70 B0WX62 A0A0N0BDM5 A0A151X135 A0A1Q3FL99
Pubmed
19121390
23622113
28756777
26354079
22118469
24495485
+ More
25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17994087 17550304 25315136 15632085 17510324 25244985 20966253 24845553 28004739 29403074 18362917 19820115 24508170 28648823 30249741 26334808 26369729 25401762 25830018 26108605 21282665 20798317
25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17994087 17550304 25315136 15632085 17510324 25244985 20966253 24845553 28004739 29403074 18362917 19820115 24508170 28648823 30249741 26334808 26369729 25401762 25830018 26108605 21282665 20798317
EMBL
BABH01007556
BABH01007557
GAIX01000062
JAA92498.1
NWSH01000568
PCG75606.1
+ More
KZ149992 PZC75539.1 KQ459597 KPI95040.1 KQ460226 KPJ16445.1 ODYU01000378 SOQ35061.1 AGBW02010703 OWR48003.1 GAMC01000733 JAC05823.1 CCAG010008775 GAKP01013713 JAC45239.1 AE014134 AAF53281.2 CM002910 KMY90071.1 CH902624 KPU74317.1 JXJN01001669 CH933807 KRG02770.1 AGB92977.1 CH940654 EDW57758.2 CH954177 EDV58701.2 CM000157 EDW88289.2 GEDC01029280 JAS08018.1 OUUW01000010 SPP85759.1 CH379062 EAL32816.3 GEBQ01014185 JAT25792.1 GFDF01011945 JAV02139.1 DS231834 EDS32623.1 CH477339 EAT43152.1 AXCN02000753 UFQS01000861 UFQT01000861 SSX07346.1 SSX27688.1 GECU01022043 GECU01006981 JAS85663.1 JAT00726.1 CH963857 KRF98387.1 APCN01004535 APCN01004536 AJWK01003455 NEVH01024950 PNF16352.1 PNF16353.1 AXCM01010803 CVRI01000020 CRK90565.1 KK853579 KDR06548.1 SPP85760.1 GEZM01013099 JAV92752.1 PYGN01000057 PSN55912.1 GEZM01013100 JAV92751.1 KQ971307 EFA11216.1 LJIG01022567 KRT79885.1 KK852476 KDR23039.1 GECZ01018693 JAS51076.1 KK107152 EZA57146.1 NNAY01001145 OXU24994.1 QOIP01000007 RLU20005.1 GBGD01002353 JAC86536.1 GDAI01001108 JAI16495.1 GBHO01035616 JAG07988.1 AJVK01002126 JR048399 AEY60653.1 GBXI01016065 JAC98226.1 LBMM01004285 KMQ92607.1 JRES01000175 KNC33606.1 GL767232 EFZ13941.1 GFDF01011234 JAV02850.1 GAKP01018234 JAC40718.1 GDHF01031887 GDHF01017679 GDHF01017273 GDHF01014108 GDHF01002433 JAI20427.1 JAI34635.1 JAI35041.1 JAI38206.1 JAI49881.1 PCG75594.1 UFQS01001311 UFQT01001311 SSX10274.1 SSX29962.1 KQ414667 KOC64771.1 GAMC01015267 GAMC01015266 JAB91288.1 UFQS01001098 UFQT01001098 SSX08996.1 SSX28907.1 GL447336 EFN86531.1 KQ434809 KZC06281.1 ACPB03024223 DS232158 EDS36323.1 KQ435863 KOX70314.1 KQ982603 KYQ53947.1 GFDL01006655 JAV28390.1
KZ149992 PZC75539.1 KQ459597 KPI95040.1 KQ460226 KPJ16445.1 ODYU01000378 SOQ35061.1 AGBW02010703 OWR48003.1 GAMC01000733 JAC05823.1 CCAG010008775 GAKP01013713 JAC45239.1 AE014134 AAF53281.2 CM002910 KMY90071.1 CH902624 KPU74317.1 JXJN01001669 CH933807 KRG02770.1 AGB92977.1 CH940654 EDW57758.2 CH954177 EDV58701.2 CM000157 EDW88289.2 GEDC01029280 JAS08018.1 OUUW01000010 SPP85759.1 CH379062 EAL32816.3 GEBQ01014185 JAT25792.1 GFDF01011945 JAV02139.1 DS231834 EDS32623.1 CH477339 EAT43152.1 AXCN02000753 UFQS01000861 UFQT01000861 SSX07346.1 SSX27688.1 GECU01022043 GECU01006981 JAS85663.1 JAT00726.1 CH963857 KRF98387.1 APCN01004535 APCN01004536 AJWK01003455 NEVH01024950 PNF16352.1 PNF16353.1 AXCM01010803 CVRI01000020 CRK90565.1 KK853579 KDR06548.1 SPP85760.1 GEZM01013099 JAV92752.1 PYGN01000057 PSN55912.1 GEZM01013100 JAV92751.1 KQ971307 EFA11216.1 LJIG01022567 KRT79885.1 KK852476 KDR23039.1 GECZ01018693 JAS51076.1 KK107152 EZA57146.1 NNAY01001145 OXU24994.1 QOIP01000007 RLU20005.1 GBGD01002353 JAC86536.1 GDAI01001108 JAI16495.1 GBHO01035616 JAG07988.1 AJVK01002126 JR048399 AEY60653.1 GBXI01016065 JAC98226.1 LBMM01004285 KMQ92607.1 JRES01000175 KNC33606.1 GL767232 EFZ13941.1 GFDF01011234 JAV02850.1 GAKP01018234 JAC40718.1 GDHF01031887 GDHF01017679 GDHF01017273 GDHF01014108 GDHF01002433 JAI20427.1 JAI34635.1 JAI35041.1 JAI38206.1 JAI49881.1 PCG75594.1 UFQS01001311 UFQT01001311 SSX10274.1 SSX29962.1 KQ414667 KOC64771.1 GAMC01015267 GAMC01015266 JAB91288.1 UFQS01001098 UFQT01001098 SSX08996.1 SSX28907.1 GL447336 EFN86531.1 KQ434809 KZC06281.1 ACPB03024223 DS232158 EDS36323.1 KQ435863 KOX70314.1 KQ982603 KYQ53947.1 GFDL01006655 JAV28390.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000092445
+ More
UP000092444 UP000091820 UP000000803 UP000007801 UP000095300 UP000092460 UP000092443 UP000078200 UP000009192 UP000008792 UP000008711 UP000192221 UP000002282 UP000268350 UP000095301 UP000001819 UP000002320 UP000008820 UP000075886 UP000075884 UP000069272 UP000075900 UP000075885 UP000075880 UP000076408 UP000075920 UP000007798 UP000075882 UP000075903 UP000076407 UP000075840 UP000092461 UP000192223 UP000235965 UP000075902 UP000075883 UP000183832 UP000027135 UP000245037 UP000007266 UP000053097 UP000215335 UP000279307 UP000005203 UP000092462 UP000036403 UP000037069 UP000053825 UP000008237 UP000076502 UP000015103 UP000053105 UP000075809
UP000092444 UP000091820 UP000000803 UP000007801 UP000095300 UP000092460 UP000092443 UP000078200 UP000009192 UP000008792 UP000008711 UP000192221 UP000002282 UP000268350 UP000095301 UP000001819 UP000002320 UP000008820 UP000075886 UP000075884 UP000069272 UP000075900 UP000075885 UP000075880 UP000076408 UP000075920 UP000007798 UP000075882 UP000075903 UP000076407 UP000075840 UP000092461 UP000192223 UP000235965 UP000075902 UP000075883 UP000183832 UP000027135 UP000245037 UP000007266 UP000053097 UP000215335 UP000279307 UP000005203 UP000092462 UP000036403 UP000037069 UP000053825 UP000008237 UP000076502 UP000015103 UP000053105 UP000075809
Interpro
IPR039860
Rost
ProteinModelPortal
H9J214
S4PPR9
A0A2A4JV12
A0A2W1BKH1
A0A194PV82
A0A0N1I933
+ More
A0A2H1V2K1 A0A212F2Q7 W8C1X8 A0A1B0A9N6 A0A1B0FBW6 A0A034VS26 A0A1A9WBX3 Q9VK03 A0A0J9R1S4 A0A0P8XHF7 A0A1I8PM89 A0A1B0AQ44 A0A1A9YBQ8 A0A1A9UJM3 A0A0Q9XDS7 M9PFU0 B4M982 B3NAY7 A0A1W4UUU2 B4P347 A0A1B6C3D6 A0A3B0KH96 A0A1I8NDS0 Q29JZ5 A0A1B6LQ50 A0A1L8D6R2 B0W430 Q17A74 A0A182QD39 A0A182NUY8 A0A182F235 A0A182RRG7 A0A336MP41 A0A182PB16 A0A182IWF9 A0A182Y5M6 A0A182WHK3 A0A1B6JNH2 A0A0Q9WQ80 A0A182L5X6 A0A182VF07 A0A182XCJ6 A0A182IDX6 A0A1B0CA98 A0A1W4WQF2 A0A2J7PJ37 A0A182TE01 A0A2J7PJ65 A0A182MUK6 A0A1J1HRB6 A0A067QSW7 A0A3B0KC43 A0A1Y1N4E4 A0A2P8ZHD3 A0A1Y1N4B9 D6W7E7 A0A0T6AYG0 A0A067RT92 A0A1B6FLK0 A0A026WND4 A0A232F2K5 A0A3L8DHS8 A0A069DS34 A0A087ZZV6 A0A0K8TRE5 A0A0A9WND3 A0A1B0CZ86 V9IHR3 A0A0A1WH74 A0A182Y5M5 A0A0J7KQI2 A0A0L0CMU8 E9IZQ9 A0A182QXJ3 A0A1L8D935 A0A034VGR5 A0A0K8V861 A0A2A4JVE8 A0A336MHR5 A0A0L7R1V2 A0A182RRG8 W8B2M2 A0A336MKK2 E2BCM1 A0A154P335 T1HR70 B0WX62 A0A0N0BDM5 A0A151X135 A0A1Q3FL99
A0A2H1V2K1 A0A212F2Q7 W8C1X8 A0A1B0A9N6 A0A1B0FBW6 A0A034VS26 A0A1A9WBX3 Q9VK03 A0A0J9R1S4 A0A0P8XHF7 A0A1I8PM89 A0A1B0AQ44 A0A1A9YBQ8 A0A1A9UJM3 A0A0Q9XDS7 M9PFU0 B4M982 B3NAY7 A0A1W4UUU2 B4P347 A0A1B6C3D6 A0A3B0KH96 A0A1I8NDS0 Q29JZ5 A0A1B6LQ50 A0A1L8D6R2 B0W430 Q17A74 A0A182QD39 A0A182NUY8 A0A182F235 A0A182RRG7 A0A336MP41 A0A182PB16 A0A182IWF9 A0A182Y5M6 A0A182WHK3 A0A1B6JNH2 A0A0Q9WQ80 A0A182L5X6 A0A182VF07 A0A182XCJ6 A0A182IDX6 A0A1B0CA98 A0A1W4WQF2 A0A2J7PJ37 A0A182TE01 A0A2J7PJ65 A0A182MUK6 A0A1J1HRB6 A0A067QSW7 A0A3B0KC43 A0A1Y1N4E4 A0A2P8ZHD3 A0A1Y1N4B9 D6W7E7 A0A0T6AYG0 A0A067RT92 A0A1B6FLK0 A0A026WND4 A0A232F2K5 A0A3L8DHS8 A0A069DS34 A0A087ZZV6 A0A0K8TRE5 A0A0A9WND3 A0A1B0CZ86 V9IHR3 A0A0A1WH74 A0A182Y5M5 A0A0J7KQI2 A0A0L0CMU8 E9IZQ9 A0A182QXJ3 A0A1L8D935 A0A034VGR5 A0A0K8V861 A0A2A4JVE8 A0A336MHR5 A0A0L7R1V2 A0A182RRG8 W8B2M2 A0A336MKK2 E2BCM1 A0A154P335 T1HR70 B0WX62 A0A0N0BDM5 A0A151X135 A0A1Q3FL99
Ontologies
PANTHER
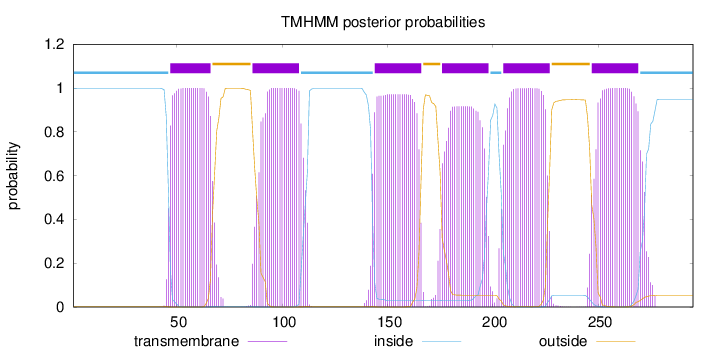
Topology
Length:
295
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
132.69935
Exp number, first 60 AAs:
14.33523
Total prob of N-in:
0.99815
POSSIBLE N-term signal
sequence
inside
1 - 46
TMhelix
47 - 66
outside
67 - 85
TMhelix
86 - 108
inside
109 - 143
TMhelix
144 - 166
outside
167 - 175
TMhelix
176 - 198
inside
199 - 204
TMhelix
205 - 227
outside
228 - 246
TMhelix
247 - 269
inside
270 - 295
Population Genetic Test Statistics
Pi
255.979031
Theta
198.535472
Tajima's D
0.859305
CLR
0.267812
CSRT
0.620668966551672
Interpretation
Uncertain
