Gene
KWMTBOMO02919
Annotation
PREDICTED:_voltage-dependent_calcium_channel_subunit_alpha-2/delta-3_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.756
Sequence
CDS
ATGCAGGCTCTGGTATATGATGCCAGAAACACAGCATGGTTCAATAAGAGTATTTCGGATTCGGCTTCCGATGAAAAAGCGTCTGAGTTCATTCAGCGTTTTGGATACATCGTGGCATTTCTGTCAACTCACAGCGGATTGACCCGATGGCAAACTCATCCGCCGAGGGAGCAGGATAACGTTAGGGCTGAATTCGGAAAGCAATGGCCCCGTTCGATAGATGAAGTGTGGTATCGACGCGCAGTCGAGCAACATTACGTGGATCCCCTCAGCTACGTTTACAGCGTTGAAATAAACACAGATAAGTTTCCGTTGAATATCAGTAATGCTGTTGTAACTGCTTCTCATGCGGTGTTCCACGGAGACGGCTACAAGAAAGCGCCAGCAGCCGTAGTCGGGTTCCAGTTCAAGCACGAACGTCTTAGCGAGTGGTTTGAGAATATAACTTCGTCGTGTGAGCACAACAAGGAATGCGTGACTTGTCTCTCGGACACCTGGGACTGTTACCTCGTCGATAACAACGGTTGGATCGTCGTCAGCGAGGATGTTACACAAAATGGCCAATTCTTTGGAAGGGCAACGACCGCCTTGCTTTGCTTTACAATGAGTGCTACGAAATATGTGCTGTAA
Protein
MQALVYDARNTAWFNKSISDSASDEKASEFIQRFGYIVAFLSTHSGLTRWQTHPPREQDNVRAEFGKQWPRSIDEVWYRRAVEQHYVDPLSYVYSVEINTDKFPLNISNAVVTASHAVFHGDGYKKAPAAVVGFQFKHERLSEWFENITSSCEHNKECVTCLSDTWDCYLVDNNGWIVVSEDVTQNGQFFGRATTALLCFTMSATKYVL
Summary
Uniprot
A0A2W1BPP2
A0A2A4JW43
A0A2A4JVU0
A0A2H1W0S0
A0A212F2P3
A0A194PP00
+ More
A0A0N1IFN0 H9J2A3 S4PWA1 A0A0L7LAP2 A0A1B0CCX0 A0A182LW46 A0A182KBT7 A0A1Y1M4X1 A0A182SSK7 Q7Q758 A0A3B0J4C4 A0A1A9YNL2 A0A3B0JNP0 B4QEH4 A0A182UDJ5 A0A139WGS5 A0A182UNW2 A0A182LJL0 A0A182PN76 A0A1S4GQ08 A0A182HM33 Q95R75 A0A182XIL0 A0A182QGK0 A0A182Y0W5 A0A084W2X5 B4HQW7 A0A182NM79 W5J6G9 A0A0Q5VMW2 A0A0R1DWW0 B4P469 A0A0Q5VMU8 A0A0R1DPV7 A0A0Q5VUW3 A0A0J9RC37 Q28XX9 A0A182J7M5 B3NRH4 N6WCF7 A0A0J9RD72 A0A182RN46 Q7K0H4 A0A0Q5VNY3 A0A0B4K7P4 A0A0Q9X2P9 B4N5V0 B4GH70 A0A1W4V4S8 A0A1W4VH47 A0A1W4VHY6 A0A182WKA1 A0A0P8ZRP9 B3MHE1 A0A0L0CQA0 A0A0P8Y6U5 A0A1B6MJM2 V9ILR0 E9IZQ6 A0A2A3EQI3 A0A3B0JFZ0 A0A1J1HY10 A0A0B5EIM5 A0A1I8QDT3 A0A154P370 A0A1I8QDV5 A0A1I8QDU9 A0A1I8QDZ2 A0A1I8QDU6 A0A0J7L4P1 E2AL65 A0A0A9X9K6 A0A0R3NPE0 N6W6G2 E2BCL9 A0A1B6IMJ4 A0A0B4K866 A0A0J9RC14 A0A151X0S3 A0A0Q9X480 A0A1W4V4S3 A0A0K8USG4 A0A1W4V4B9 A0A0K8VZ34 A0A0Q9W3E8 A0A1W4VGK1 A0A0Q9WEY5 A0A0Q9W3C2 A0A0Q9WEB3 A0A1W4VH43 A0A0Q9W3Q6 A0A0K8U1W5 A0A0Q9WF22 B0WES9
A0A0N1IFN0 H9J2A3 S4PWA1 A0A0L7LAP2 A0A1B0CCX0 A0A182LW46 A0A182KBT7 A0A1Y1M4X1 A0A182SSK7 Q7Q758 A0A3B0J4C4 A0A1A9YNL2 A0A3B0JNP0 B4QEH4 A0A182UDJ5 A0A139WGS5 A0A182UNW2 A0A182LJL0 A0A182PN76 A0A1S4GQ08 A0A182HM33 Q95R75 A0A182XIL0 A0A182QGK0 A0A182Y0W5 A0A084W2X5 B4HQW7 A0A182NM79 W5J6G9 A0A0Q5VMW2 A0A0R1DWW0 B4P469 A0A0Q5VMU8 A0A0R1DPV7 A0A0Q5VUW3 A0A0J9RC37 Q28XX9 A0A182J7M5 B3NRH4 N6WCF7 A0A0J9RD72 A0A182RN46 Q7K0H4 A0A0Q5VNY3 A0A0B4K7P4 A0A0Q9X2P9 B4N5V0 B4GH70 A0A1W4V4S8 A0A1W4VH47 A0A1W4VHY6 A0A182WKA1 A0A0P8ZRP9 B3MHE1 A0A0L0CQA0 A0A0P8Y6U5 A0A1B6MJM2 V9ILR0 E9IZQ6 A0A2A3EQI3 A0A3B0JFZ0 A0A1J1HY10 A0A0B5EIM5 A0A1I8QDT3 A0A154P370 A0A1I8QDV5 A0A1I8QDU9 A0A1I8QDZ2 A0A1I8QDU6 A0A0J7L4P1 E2AL65 A0A0A9X9K6 A0A0R3NPE0 N6W6G2 E2BCL9 A0A1B6IMJ4 A0A0B4K866 A0A0J9RC14 A0A151X0S3 A0A0Q9X480 A0A1W4V4S3 A0A0K8USG4 A0A1W4V4B9 A0A0K8VZ34 A0A0Q9W3E8 A0A1W4VGK1 A0A0Q9WEY5 A0A0Q9W3C2 A0A0Q9WEB3 A0A1W4VH43 A0A0Q9W3Q6 A0A0K8U1W5 A0A0Q9WF22 B0WES9
Pubmed
28756777
22118469
26354079
19121390
23622113
26227816
+ More
28004739 12364791 17994087 18362917 19820115 20966253 25244985 24438588 20920257 23761445 17550304 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 21282665 20798317 25401762
28004739 12364791 17994087 18362917 19820115 20966253 25244985 24438588 20920257 23761445 17550304 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 21282665 20798317 25401762
EMBL
KZ149992
PZC75535.1
NWSH01000568
PCG75602.1
PCG75603.1
ODYU01005348
+ More
SOQ46104.1 AGBW02010703 OWR48004.1 KQ459597 KPI95037.1 KQ460226 KPJ16443.1 BABH01007564 BABH01007565 GAIX01007148 JAA85412.1 JTDY01001946 KOB72469.1 AJWK01007163 AJWK01007164 AXCM01011171 GEZM01041950 JAV80088.1 AAAB01008960 EAA10906.4 OUUW01000001 SPP74262.1 SPP74261.1 CM000362 EDX06964.1 KQ971345 KYB27055.1 APCN01004331 APCN01004332 AY061583 AAL29131.1 AXCN02000940 ATLV01019720 KE525278 KFB44569.1 CH480816 EDW47764.1 ADMH02002072 ETN59596.1 CH954179 KQS62710.1 CM000158 KRJ99302.1 EDW90579.2 KQS62711.1 KRJ99303.1 KQS62709.1 CM002911 KMY93563.1 CM000071 EAL26187.3 EDV56126.2 ENO01819.1 KMY93564.1 AE013599 AY069830 AAF58335.2 AAL39975.1 KQS62708.1 AFH08058.1 CH964154 KRF99110.1 EDW79739.1 CH479183 EDW35840.1 CH902619 KPU77119.1 EDV37941.1 JRES01000080 KNC34362.1 KPU77120.1 GEBQ01003928 JAT36049.1 JR051907 AEY61587.1 GL767232 EFZ13976.1 KZ288194 PBC33967.1 SPP74260.1 CVRI01000036 CRK92984.1 KJ485707 AJE70859.1 KQ434809 KZC06283.1 LBMM01000796 KMQ97474.1 GL440492 EFN65820.1 GBHO01028111 JAG15493.1 KRT02903.1 ENO01818.1 GL447336 EFN86529.1 GECU01019559 JAS88147.1 AFH08057.1 KMY93562.1 KQ982603 KYQ53949.1 KRF99111.1 GDHF01022793 JAI29521.1 GDHF01008205 JAI44109.1 CH940648 KRF79596.1 KRF79595.1 KRF79594.1 KRF79599.1 KRF79597.1 GDHF01031612 GDHF01016000 JAI20702.1 JAI36314.1 KRF79598.1 DS231911 EDS45842.1
SOQ46104.1 AGBW02010703 OWR48004.1 KQ459597 KPI95037.1 KQ460226 KPJ16443.1 BABH01007564 BABH01007565 GAIX01007148 JAA85412.1 JTDY01001946 KOB72469.1 AJWK01007163 AJWK01007164 AXCM01011171 GEZM01041950 JAV80088.1 AAAB01008960 EAA10906.4 OUUW01000001 SPP74262.1 SPP74261.1 CM000362 EDX06964.1 KQ971345 KYB27055.1 APCN01004331 APCN01004332 AY061583 AAL29131.1 AXCN02000940 ATLV01019720 KE525278 KFB44569.1 CH480816 EDW47764.1 ADMH02002072 ETN59596.1 CH954179 KQS62710.1 CM000158 KRJ99302.1 EDW90579.2 KQS62711.1 KRJ99303.1 KQS62709.1 CM002911 KMY93563.1 CM000071 EAL26187.3 EDV56126.2 ENO01819.1 KMY93564.1 AE013599 AY069830 AAF58335.2 AAL39975.1 KQS62708.1 AFH08058.1 CH964154 KRF99110.1 EDW79739.1 CH479183 EDW35840.1 CH902619 KPU77119.1 EDV37941.1 JRES01000080 KNC34362.1 KPU77120.1 GEBQ01003928 JAT36049.1 JR051907 AEY61587.1 GL767232 EFZ13976.1 KZ288194 PBC33967.1 SPP74260.1 CVRI01000036 CRK92984.1 KJ485707 AJE70859.1 KQ434809 KZC06283.1 LBMM01000796 KMQ97474.1 GL440492 EFN65820.1 GBHO01028111 JAG15493.1 KRT02903.1 ENO01818.1 GL447336 EFN86529.1 GECU01019559 JAS88147.1 AFH08057.1 KMY93562.1 KQ982603 KYQ53949.1 KRF99111.1 GDHF01022793 JAI29521.1 GDHF01008205 JAI44109.1 CH940648 KRF79596.1 KRF79595.1 KRF79594.1 KRF79599.1 KRF79597.1 GDHF01031612 GDHF01016000 JAI20702.1 JAI36314.1 KRF79598.1 DS231911 EDS45842.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000005204
UP000037510
+ More
UP000092461 UP000075883 UP000075881 UP000075901 UP000007062 UP000268350 UP000092443 UP000000304 UP000075902 UP000007266 UP000075903 UP000075882 UP000075885 UP000075840 UP000076407 UP000075886 UP000076408 UP000030765 UP000001292 UP000075884 UP000000673 UP000008711 UP000002282 UP000001819 UP000075880 UP000075900 UP000000803 UP000007798 UP000008744 UP000192221 UP000075920 UP000007801 UP000037069 UP000242457 UP000183832 UP000095300 UP000076502 UP000036403 UP000000311 UP000008237 UP000075809 UP000008792 UP000002320
UP000092461 UP000075883 UP000075881 UP000075901 UP000007062 UP000268350 UP000092443 UP000000304 UP000075902 UP000007266 UP000075903 UP000075882 UP000075885 UP000075840 UP000076407 UP000075886 UP000076408 UP000030765 UP000001292 UP000075884 UP000000673 UP000008711 UP000002282 UP000001819 UP000075880 UP000075900 UP000000803 UP000007798 UP000008744 UP000192221 UP000075920 UP000007801 UP000037069 UP000242457 UP000183832 UP000095300 UP000076502 UP000036403 UP000000311 UP000008237 UP000075809 UP000008792 UP000002320
Pfam
Interpro
SUPFAM
SSF53300
SSF53300
Gene 3D
ProteinModelPortal
A0A2W1BPP2
A0A2A4JW43
A0A2A4JVU0
A0A2H1W0S0
A0A212F2P3
A0A194PP00
+ More
A0A0N1IFN0 H9J2A3 S4PWA1 A0A0L7LAP2 A0A1B0CCX0 A0A182LW46 A0A182KBT7 A0A1Y1M4X1 A0A182SSK7 Q7Q758 A0A3B0J4C4 A0A1A9YNL2 A0A3B0JNP0 B4QEH4 A0A182UDJ5 A0A139WGS5 A0A182UNW2 A0A182LJL0 A0A182PN76 A0A1S4GQ08 A0A182HM33 Q95R75 A0A182XIL0 A0A182QGK0 A0A182Y0W5 A0A084W2X5 B4HQW7 A0A182NM79 W5J6G9 A0A0Q5VMW2 A0A0R1DWW0 B4P469 A0A0Q5VMU8 A0A0R1DPV7 A0A0Q5VUW3 A0A0J9RC37 Q28XX9 A0A182J7M5 B3NRH4 N6WCF7 A0A0J9RD72 A0A182RN46 Q7K0H4 A0A0Q5VNY3 A0A0B4K7P4 A0A0Q9X2P9 B4N5V0 B4GH70 A0A1W4V4S8 A0A1W4VH47 A0A1W4VHY6 A0A182WKA1 A0A0P8ZRP9 B3MHE1 A0A0L0CQA0 A0A0P8Y6U5 A0A1B6MJM2 V9ILR0 E9IZQ6 A0A2A3EQI3 A0A3B0JFZ0 A0A1J1HY10 A0A0B5EIM5 A0A1I8QDT3 A0A154P370 A0A1I8QDV5 A0A1I8QDU9 A0A1I8QDZ2 A0A1I8QDU6 A0A0J7L4P1 E2AL65 A0A0A9X9K6 A0A0R3NPE0 N6W6G2 E2BCL9 A0A1B6IMJ4 A0A0B4K866 A0A0J9RC14 A0A151X0S3 A0A0Q9X480 A0A1W4V4S3 A0A0K8USG4 A0A1W4V4B9 A0A0K8VZ34 A0A0Q9W3E8 A0A1W4VGK1 A0A0Q9WEY5 A0A0Q9W3C2 A0A0Q9WEB3 A0A1W4VH43 A0A0Q9W3Q6 A0A0K8U1W5 A0A0Q9WF22 B0WES9
A0A0N1IFN0 H9J2A3 S4PWA1 A0A0L7LAP2 A0A1B0CCX0 A0A182LW46 A0A182KBT7 A0A1Y1M4X1 A0A182SSK7 Q7Q758 A0A3B0J4C4 A0A1A9YNL2 A0A3B0JNP0 B4QEH4 A0A182UDJ5 A0A139WGS5 A0A182UNW2 A0A182LJL0 A0A182PN76 A0A1S4GQ08 A0A182HM33 Q95R75 A0A182XIL0 A0A182QGK0 A0A182Y0W5 A0A084W2X5 B4HQW7 A0A182NM79 W5J6G9 A0A0Q5VMW2 A0A0R1DWW0 B4P469 A0A0Q5VMU8 A0A0R1DPV7 A0A0Q5VUW3 A0A0J9RC37 Q28XX9 A0A182J7M5 B3NRH4 N6WCF7 A0A0J9RD72 A0A182RN46 Q7K0H4 A0A0Q5VNY3 A0A0B4K7P4 A0A0Q9X2P9 B4N5V0 B4GH70 A0A1W4V4S8 A0A1W4VH47 A0A1W4VHY6 A0A182WKA1 A0A0P8ZRP9 B3MHE1 A0A0L0CQA0 A0A0P8Y6U5 A0A1B6MJM2 V9ILR0 E9IZQ6 A0A2A3EQI3 A0A3B0JFZ0 A0A1J1HY10 A0A0B5EIM5 A0A1I8QDT3 A0A154P370 A0A1I8QDV5 A0A1I8QDU9 A0A1I8QDZ2 A0A1I8QDU6 A0A0J7L4P1 E2AL65 A0A0A9X9K6 A0A0R3NPE0 N6W6G2 E2BCL9 A0A1B6IMJ4 A0A0B4K866 A0A0J9RC14 A0A151X0S3 A0A0Q9X480 A0A1W4V4S3 A0A0K8USG4 A0A1W4V4B9 A0A0K8VZ34 A0A0Q9W3E8 A0A1W4VGK1 A0A0Q9WEY5 A0A0Q9W3C2 A0A0Q9WEB3 A0A1W4VH43 A0A0Q9W3Q6 A0A0K8U1W5 A0A0Q9WF22 B0WES9
Ontologies
GO
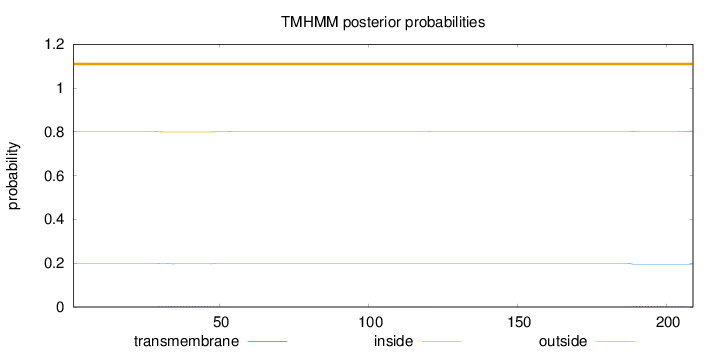
Topology
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14179
Exp number, first 60 AAs:
0.0637
Total prob of N-in:
0.19857
outside
1 - 209
Population Genetic Test Statistics
Pi
263.528947
Theta
183.052307
Tajima's D
2.315646
CLR
0.380179
CSRT
0.925153742312884
Interpretation
Uncertain
