Pre Gene Modal
BGIBMGA003549
Annotation
mitochondrial_translational_release_factor_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.294 Mitochondrial Reliability : 1.465 Nuclear Reliability : 1.492
Sequence
CDS
ATGTCGTTTCCGAAACTTCTCTTTCTTCGCTCGTTCTTGAATTCTGAAAGACAATTATTGCAATTTCGTAGACACTGCTCACATAATATCATGAACCTTTCGGACCCTGCTATTCAAACATATCTTAAGCATTTGATGGTAGAGCATGAAAACTTGCTAAGTAAAATGAAAAAATCCTCCGAAGAAGCTAGACGCTTACAGGAAATAAAACCAATCGTAAATGTCCTAGAACAAAGGATAGCTTTATATGATAGTATTGAATCACTTAAAGAATTAAATAAAAAAGAAGGTGACGATAGTGAGATAAAAAAGATGATAAAGGAAGAAGCGGCGATATATCTGAAACGTCTAAAGGAAATAGATGACGATTTACAGTTAATTCTTCTCGAACCACCATTAACTGAAGGTGGTGTACTCTTAGAACTAACTGCAGGAGCTGGAGGGCAAGAAGCGATGTTATTTGCAAAAGAACTCTTCAACATGTACCAGTCTTATGCTATCAACAAAGGTTGGGAAGTTGATATAGCATCATTTGACTTAACCGAAATAGGTGGAGTTCGTAAAGGATCAATGTTGATACAAGGTTTTGGTGTACCTGAACTGATGAGGATGGAGGCTGGAGTTCATCGTGTTCAAAGAATTCCTGCTACGGAGAAAGGTGGACGTATACATACTAGTACAGTCACTGTCGCTATTTTACCACAGCCCTCTGAAATAGAACTGAACATCCCTGAACGTGATGTAGTAATAGAGACAAAAAGAGCTAGTGGGGCTGGAGGTCAGCATGTAAACACAACCGATAGTGCTGTAAGGCTAATACATACACCGACGGGAACAACGGTTGAGTGTCAAGAAGGAAGATCACAAATAAAGAACAAACAAATTGCTATGCAGAAACTGAGAACACTTTTATTGGAGAAGCAGATACAAGAACAAGCTTTAAAACTGCAAAGTGAGAGGAAATCGCAGGTTGGGTCTGGCAACAGAAATGAAAAAATAAGAACATACAACTTCCCTCAAGATAGAGTGACAGAACATCGAGATGGTGGTGGGACATTCCATAATCTCAAAGGTTTTATGGAAGGTGGAGAGCAACTAGAACAGTTACAGGAGTCACTCCTTAAGAAAGAAAGACACAAGCTTTTACTGGAAGAAATTGAAAACATTCTGAATAAACACAGAGGAAAAACCATTGAAAGTTCCACCAGTCATTAA
Protein
MSFPKLLFLRSFLNSERQLLQFRRHCSHNIMNLSDPAIQTYLKHLMVEHENLLSKMKKSSEEARRLQEIKPIVNVLEQRIALYDSIESLKELNKKEGDDSEIKKMIKEEAAIYLKRLKEIDDDLQLILLEPPLTEGGVLLELTAGAGGQEAMLFAKELFNMYQSYAINKGWEVDIASFDLTEIGGVRKGSMLIQGFGVPELMRMEAGVHRVQRIPATEKGGRIHTSTVTVAILPQPSEIELNIPERDVVIETKRASGAGGQHVNTTDSAVRLIHTPTGTTVECQEGRSQIKNKQIAMQKLRTLLLEKQIQEQALKLQSERKSQVGSGNRNEKIRTYNFPQDRVTEHRDGGGTFHNLKGFMEGGEQLEQLQESLLKKERHKLLLEEIENILNKHRGKTIESSTSH
Summary
Uniprot
Q1HPR9
H9J212
A0A0L7L6E3
A0A2A4JW28
A0A194PNM7
A0A0N1PJH0
+ More
A0A2H1VZ72 A0A212F2N3 A0A2W1BUV1 A0A2W1BKC6 A0A336LF00 A0A336N0N0 W5JEC4 A0A139WI12 A0A2M4BQ23 A0A2M4BPM3 A0A2J7PJ26 A0A139WHU5 A0A182FTW3 A0A2M4ALE6 A0A067QFM0 A0A182VAL3 Q7PZU0 A0A182WD14 A0A182HPJ2 A0A182THH1 A0A182MQU2 Q29KH7 A0A182JUN5 B4JCT3 A0A182YCB0 A0A3B0JZ81 A0A0L0BZ12 A0A1Q3F5W8 B0XII8 A0A1W4V7W5 B3MN37 A0A2P8YFP1 J3JXQ3 N6T309 U4UHQ1 A0A1Y1K8Y0 B3N385 A0A084WI94 Q16KB5 Q16L13 A0A0J9TMJ3 Q9VK20 A0A182P1D5 B4P2N5 A0A182G1Q2 A0A023ET49 A0A182XR80 B4M9A2 W8BSI3 A0A182KTC3 A0A1I8NB30 B4KHM1 A0A0M4E8N5 A0A0A1X0P6 A0A0P4VWY9 A0A182JG15 A0A182QLP3 A0A1I8P5D6 A0A1B6C7R2 A0A1W4WHC9 A0A034WSW3 B4MX15 A0A026WMA9 A0A1A9UV03 A0A2M3ZJG0 A0A2M3ZJE3 A0A0K8UJT2 A0A1B0ASY6 A0A1A9XYD2 A0A1B0GJ78 A0A1L8DRV8 A0A2A3ERX4 A0A0A9Y3A0 A0A1L8DRR7 A0A087ZZV4 A0A2R7WD06 A0A146LJL5 A0A3B3QDC6 A0A0C9R1C1 A0A0M8ZVE0
A0A2H1VZ72 A0A212F2N3 A0A2W1BUV1 A0A2W1BKC6 A0A336LF00 A0A336N0N0 W5JEC4 A0A139WI12 A0A2M4BQ23 A0A2M4BPM3 A0A2J7PJ26 A0A139WHU5 A0A182FTW3 A0A2M4ALE6 A0A067QFM0 A0A182VAL3 Q7PZU0 A0A182WD14 A0A182HPJ2 A0A182THH1 A0A182MQU2 Q29KH7 A0A182JUN5 B4JCT3 A0A182YCB0 A0A3B0JZ81 A0A0L0BZ12 A0A1Q3F5W8 B0XII8 A0A1W4V7W5 B3MN37 A0A2P8YFP1 J3JXQ3 N6T309 U4UHQ1 A0A1Y1K8Y0 B3N385 A0A084WI94 Q16KB5 Q16L13 A0A0J9TMJ3 Q9VK20 A0A182P1D5 B4P2N5 A0A182G1Q2 A0A023ET49 A0A182XR80 B4M9A2 W8BSI3 A0A182KTC3 A0A1I8NB30 B4KHM1 A0A0M4E8N5 A0A0A1X0P6 A0A0P4VWY9 A0A182JG15 A0A182QLP3 A0A1I8P5D6 A0A1B6C7R2 A0A1W4WHC9 A0A034WSW3 B4MX15 A0A026WMA9 A0A1A9UV03 A0A2M3ZJG0 A0A2M3ZJE3 A0A0K8UJT2 A0A1B0ASY6 A0A1A9XYD2 A0A1B0GJ78 A0A1L8DRV8 A0A2A3ERX4 A0A0A9Y3A0 A0A1L8DRR7 A0A087ZZV4 A0A2R7WD06 A0A146LJL5 A0A3B3QDC6 A0A0C9R1C1 A0A0M8ZVE0
Pubmed
19121390
26227816
26354079
22118469
28756777
20920257
+ More
23761445 18362917 19820115 24845553 12364791 15632085 17994087 25244985 26108605 29403074 22516182 23537049 28004739 24438588 17510324 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26483478 24945155 24495485 20966253 25315136 25830018 25348373 24508170 30249741 25401762 26823975 29240929
23761445 18362917 19820115 24845553 12364791 15632085 17994087 25244985 26108605 29403074 22516182 23537049 28004739 24438588 17510324 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26483478 24945155 24495485 20966253 25315136 25830018 25348373 24508170 30249741 25401762 26823975 29240929
EMBL
DQ443333
ABF51422.1
BABH01007566
JTDY01002668
KOB70975.1
NWSH01000568
+ More
PCG75600.1 KQ459597 KPI95036.1 KQ460226 KPJ16442.1 ODYU01005348 SOQ46103.1 AGBW02010703 OWR48005.1 KZ149992 PZC75533.1 PZC75532.1 UFQS01004024 UFQT01004024 SSX16109.1 SSX35438.1 UFQS01004067 UFQT01004067 SSX16144.1 SSX35471.1 ADMH02001378 ETN62707.1 KQ971342 KYB27534.1 GGFJ01005880 MBW55021.1 GGFJ01005879 MBW55020.1 NEVH01024950 PNF16345.1 KYB27532.1 GGFK01008263 MBW41584.1 KK853579 KDR06565.1 AAAB01008986 EAA00433.4 APCN01003261 AXCM01003529 CH379061 EAL33198.1 CH916368 EDW03172.1 OUUW01000010 SPP85732.1 JRES01001125 KNC25292.1 GFDL01012098 JAV22947.1 DS233313 EDS29387.1 CH902620 EDV32015.1 PYGN01000632 PSN43069.1 BT128024 AEE62985.1 APGK01054225 KB741248 ENN71938.1 KB632305 ERL91863.1 GEZM01088917 GEZM01088916 GEZM01088913 JAV57834.1 CH954177 EDV58725.1 ATLV01023923 KE525347 KFB49938.1 CH477969 EAT34741.1 CH477928 EAT34990.1 CM002910 KMY90045.1 AE014134 AY061245 AAF53262.1 AAL28793.1 CM000157 EDW88265.1 JXUM01039007 KQ561187 KXJ79426.1 GAPW01001612 JAC11986.1 CH940654 EDW57778.1 GAMC01004463 JAC02093.1 CH933807 EDW12300.1 CP012523 ALC38401.1 GBXI01010079 JAD04213.1 GDRN01101499 JAI58359.1 AXCN02001102 GEDC01027751 JAS09547.1 GAKP01001692 JAC57260.1 CH963857 EDW76654.1 KK107152 QOIP01000007 EZA57150.1 RLU20009.1 GGFM01007902 MBW28653.1 GGFM01007878 MBW28629.1 GDHF01025513 JAI26801.1 JXJN01003038 AJWK01019500 GFDF01005019 JAV09065.1 KZ288194 PBC33966.1 GBHO01016980 GBRD01008056 JAG26624.1 JAG57765.1 GFDF01005020 JAV09064.1 KK854638 PTY17554.1 GDHC01011553 JAQ07076.1 GBYB01001755 JAG71522.1 KQ435863 KOX70319.1
PCG75600.1 KQ459597 KPI95036.1 KQ460226 KPJ16442.1 ODYU01005348 SOQ46103.1 AGBW02010703 OWR48005.1 KZ149992 PZC75533.1 PZC75532.1 UFQS01004024 UFQT01004024 SSX16109.1 SSX35438.1 UFQS01004067 UFQT01004067 SSX16144.1 SSX35471.1 ADMH02001378 ETN62707.1 KQ971342 KYB27534.1 GGFJ01005880 MBW55021.1 GGFJ01005879 MBW55020.1 NEVH01024950 PNF16345.1 KYB27532.1 GGFK01008263 MBW41584.1 KK853579 KDR06565.1 AAAB01008986 EAA00433.4 APCN01003261 AXCM01003529 CH379061 EAL33198.1 CH916368 EDW03172.1 OUUW01000010 SPP85732.1 JRES01001125 KNC25292.1 GFDL01012098 JAV22947.1 DS233313 EDS29387.1 CH902620 EDV32015.1 PYGN01000632 PSN43069.1 BT128024 AEE62985.1 APGK01054225 KB741248 ENN71938.1 KB632305 ERL91863.1 GEZM01088917 GEZM01088916 GEZM01088913 JAV57834.1 CH954177 EDV58725.1 ATLV01023923 KE525347 KFB49938.1 CH477969 EAT34741.1 CH477928 EAT34990.1 CM002910 KMY90045.1 AE014134 AY061245 AAF53262.1 AAL28793.1 CM000157 EDW88265.1 JXUM01039007 KQ561187 KXJ79426.1 GAPW01001612 JAC11986.1 CH940654 EDW57778.1 GAMC01004463 JAC02093.1 CH933807 EDW12300.1 CP012523 ALC38401.1 GBXI01010079 JAD04213.1 GDRN01101499 JAI58359.1 AXCN02001102 GEDC01027751 JAS09547.1 GAKP01001692 JAC57260.1 CH963857 EDW76654.1 KK107152 QOIP01000007 EZA57150.1 RLU20009.1 GGFM01007902 MBW28653.1 GGFM01007878 MBW28629.1 GDHF01025513 JAI26801.1 JXJN01003038 AJWK01019500 GFDF01005019 JAV09065.1 KZ288194 PBC33966.1 GBHO01016980 GBRD01008056 JAG26624.1 JAG57765.1 GFDF01005020 JAV09064.1 KK854638 PTY17554.1 GDHC01011553 JAQ07076.1 GBYB01001755 JAG71522.1 KQ435863 KOX70319.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000000673 UP000007266 UP000235965 UP000069272 UP000027135 UP000075903 UP000007062 UP000075920 UP000075840 UP000075902 UP000075883 UP000001819 UP000075881 UP000001070 UP000076408 UP000268350 UP000037069 UP000002320 UP000192221 UP000007801 UP000245037 UP000019118 UP000030742 UP000008711 UP000030765 UP000008820 UP000000803 UP000075885 UP000002282 UP000069940 UP000249989 UP000076407 UP000008792 UP000075882 UP000095301 UP000009192 UP000092553 UP000075880 UP000075886 UP000095300 UP000192223 UP000007798 UP000053097 UP000279307 UP000078200 UP000092460 UP000092443 UP000092461 UP000242457 UP000005203 UP000261540 UP000053105
UP000000673 UP000007266 UP000235965 UP000069272 UP000027135 UP000075903 UP000007062 UP000075920 UP000075840 UP000075902 UP000075883 UP000001819 UP000075881 UP000001070 UP000076408 UP000268350 UP000037069 UP000002320 UP000192221 UP000007801 UP000245037 UP000019118 UP000030742 UP000008711 UP000030765 UP000008820 UP000000803 UP000075885 UP000002282 UP000069940 UP000249989 UP000076407 UP000008792 UP000075882 UP000095301 UP000009192 UP000092553 UP000075880 UP000075886 UP000095300 UP000192223 UP000007798 UP000053097 UP000279307 UP000078200 UP000092460 UP000092443 UP000092461 UP000242457 UP000005203 UP000261540 UP000053105
ProteinModelPortal
Q1HPR9
H9J212
A0A0L7L6E3
A0A2A4JW28
A0A194PNM7
A0A0N1PJH0
+ More
A0A2H1VZ72 A0A212F2N3 A0A2W1BUV1 A0A2W1BKC6 A0A336LF00 A0A336N0N0 W5JEC4 A0A139WI12 A0A2M4BQ23 A0A2M4BPM3 A0A2J7PJ26 A0A139WHU5 A0A182FTW3 A0A2M4ALE6 A0A067QFM0 A0A182VAL3 Q7PZU0 A0A182WD14 A0A182HPJ2 A0A182THH1 A0A182MQU2 Q29KH7 A0A182JUN5 B4JCT3 A0A182YCB0 A0A3B0JZ81 A0A0L0BZ12 A0A1Q3F5W8 B0XII8 A0A1W4V7W5 B3MN37 A0A2P8YFP1 J3JXQ3 N6T309 U4UHQ1 A0A1Y1K8Y0 B3N385 A0A084WI94 Q16KB5 Q16L13 A0A0J9TMJ3 Q9VK20 A0A182P1D5 B4P2N5 A0A182G1Q2 A0A023ET49 A0A182XR80 B4M9A2 W8BSI3 A0A182KTC3 A0A1I8NB30 B4KHM1 A0A0M4E8N5 A0A0A1X0P6 A0A0P4VWY9 A0A182JG15 A0A182QLP3 A0A1I8P5D6 A0A1B6C7R2 A0A1W4WHC9 A0A034WSW3 B4MX15 A0A026WMA9 A0A1A9UV03 A0A2M3ZJG0 A0A2M3ZJE3 A0A0K8UJT2 A0A1B0ASY6 A0A1A9XYD2 A0A1B0GJ78 A0A1L8DRV8 A0A2A3ERX4 A0A0A9Y3A0 A0A1L8DRR7 A0A087ZZV4 A0A2R7WD06 A0A146LJL5 A0A3B3QDC6 A0A0C9R1C1 A0A0M8ZVE0
A0A2H1VZ72 A0A212F2N3 A0A2W1BUV1 A0A2W1BKC6 A0A336LF00 A0A336N0N0 W5JEC4 A0A139WI12 A0A2M4BQ23 A0A2M4BPM3 A0A2J7PJ26 A0A139WHU5 A0A182FTW3 A0A2M4ALE6 A0A067QFM0 A0A182VAL3 Q7PZU0 A0A182WD14 A0A182HPJ2 A0A182THH1 A0A182MQU2 Q29KH7 A0A182JUN5 B4JCT3 A0A182YCB0 A0A3B0JZ81 A0A0L0BZ12 A0A1Q3F5W8 B0XII8 A0A1W4V7W5 B3MN37 A0A2P8YFP1 J3JXQ3 N6T309 U4UHQ1 A0A1Y1K8Y0 B3N385 A0A084WI94 Q16KB5 Q16L13 A0A0J9TMJ3 Q9VK20 A0A182P1D5 B4P2N5 A0A182G1Q2 A0A023ET49 A0A182XR80 B4M9A2 W8BSI3 A0A182KTC3 A0A1I8NB30 B4KHM1 A0A0M4E8N5 A0A0A1X0P6 A0A0P4VWY9 A0A182JG15 A0A182QLP3 A0A1I8P5D6 A0A1B6C7R2 A0A1W4WHC9 A0A034WSW3 B4MX15 A0A026WMA9 A0A1A9UV03 A0A2M3ZJG0 A0A2M3ZJE3 A0A0K8UJT2 A0A1B0ASY6 A0A1A9XYD2 A0A1B0GJ78 A0A1L8DRV8 A0A2A3ERX4 A0A0A9Y3A0 A0A1L8DRR7 A0A087ZZV4 A0A2R7WD06 A0A146LJL5 A0A3B3QDC6 A0A0C9R1C1 A0A0M8ZVE0
PDB
4V7P
E-value=1.59206e-56,
Score=555
Ontologies
KEGG
GO
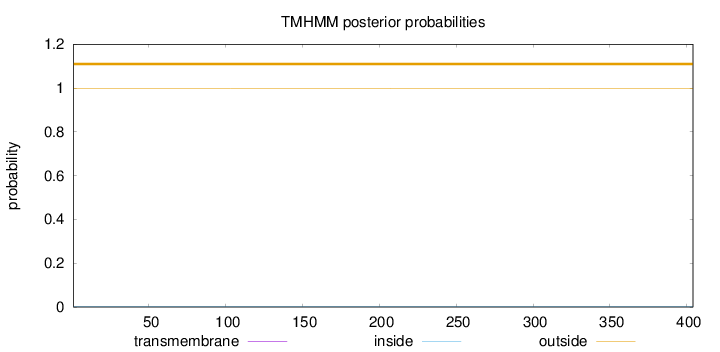
Topology
Length:
404
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00047
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00074
outside
1 - 404
Population Genetic Test Statistics
Pi
257.149031
Theta
176.761288
Tajima's D
1.435176
CLR
0.514402
CSRT
0.769411529423529
Interpretation
Uncertain
