Gene
KWMTBOMO02915
Pre Gene Modal
BGIBMGA003547
Annotation
Fork_head_domain-containing_protein_FD4_[Operophtera_brumata]
Transcription factor
Location in the cell
Nuclear Reliability : 3.762
Sequence
CDS
ATGCCGCGACCTTCCCGCGATTCTTATGGTGATCAGAAGCCACCTTACTCATACATTTCTTTGACCGCGATGGCGATCTGGAGTTCCCCGGAGCGTATGCTGCCGCTGTCGGAGATCTACCGGTTCATTACGGATCGGTTCCCGTACTACCGGCGCAACACGCAGCGCTGGCAGAACTCGCTGCGACACAACCTGTCCTTCAACGACTGCTTCGTGAAGGTGCCGCGCCGCCCCGACCGGCCCGGCAAGGGCGCGTACTGGACCTTGCATCCCCAGGCGTTTGACATGTTCGAAAATGGCTCTTTGCTTCGTCGGCGGAAACGTTTTAAGTTACAAAAGGGAGAAAAAGATAACTTGAATGCTGAATTAGCGGCTTTGGCGAATTTTAATCGGGCGTTTTTAGCCCGACAAGCGGCCGGGCCGCCGCCGCCCGCGAGTATACCCACAGCGACTTTGTACACATCAACGTTGTGCTCTCAACTAAGCCCAGAGCCGGCAGAGCTTCCTGACGTGACTCCTATCACTATGTTGCCCAGGGAAAGACCGCGGAGGGCGTTTACGATCGATGCGTTATTGGAGCCAGATTCGCCTCGCTTGTCACCGTCGCCGCCACAGCCCATGATCAGCCTCGAGGCTCGCATGGCGGCTCCGTACGTCCTGGCCGCGCAACGCTACCACGCCGAGCTGCTGCAGGCCGCGGCGCACGCGCTTCGACCCTGCTATTTACCGCCGCCACCTTTACCGGTCGCGTGA
Protein
MPRPSRDSYGDQKPPYSYISLTAMAIWSSPERMLPLSEIYRFITDRFPYYRRNTQRWQNSLRHNLSFNDCFVKVPRRPDRPGKGAYWTLHPQAFDMFENGSLLRRRKRFKLQKGEKDNLNAELAALANFNRAFLARQAAGPPPPASIPTATLYTSTLCSQLSPEPAELPDVTPITMLPRERPRRAFTIDALLEPDSPRLSPSPPQPMISLEARMAAPYVLAAQRYHAELLQAAAHALRPCYLPPPPLPVA
Summary
Uniprot
H9J210
A0A0L7LSA3
A0A2H1V4U3
A0A0N1IAY8
A0A2A4J7M3
A0A2W1B6E1
+ More
A0A194PQL6 A0A212F2P8 A0A2A4JW33 A0A2W1BML0 A0A212F2P9 A0A0N0PDE0 A0A2H1V3J5 A0A194PV77 A0A0L7LSZ6 A0A0N0PDS3 A0A194PPY8 B0X157 A0A2A4J7E0 A0A2W1BN29 A0A2J7R3D1 A0A067RL19 A0A182GQV0 A0A182H7J4 A0A182TKZ5 A0A1B0GME1 Q17GJ8 A0A182V4W8 A0A2P8YVZ1 A0A1A9W2Q1 A0A1A9Y7F8 A0A1B0BTB3 A0A182LF41 A0A1W4WPX5 B4NJV5 A0A1A9VM15 A0A1B0AHF0 A0A1B0GDY2 B4GE28 D6W6W8 A0A182RA03 A0A182N5T6 A0A084WRG2 A0A182QTF8 N6UJA5 A0A182PAE0 B3M1Z1 B4KBG6 B3P6S4 F5HK36 A0A182XI20 A0A182HT03 A0A182XWX6 A0A182SFZ9 A0A182LVV2 A0A182VXD0 A0A2R7WXR3 A0A1J1HKM7 A0A182FT34 A0A182JBJ8 E0VH79 A0A182G9Q1 Q1DGX0
A0A194PQL6 A0A212F2P8 A0A2A4JW33 A0A2W1BML0 A0A212F2P9 A0A0N0PDE0 A0A2H1V3J5 A0A194PV77 A0A0L7LSZ6 A0A0N0PDS3 A0A194PPY8 B0X157 A0A2A4J7E0 A0A2W1BN29 A0A2J7R3D1 A0A067RL19 A0A182GQV0 A0A182H7J4 A0A182TKZ5 A0A1B0GME1 Q17GJ8 A0A182V4W8 A0A2P8YVZ1 A0A1A9W2Q1 A0A1A9Y7F8 A0A1B0BTB3 A0A182LF41 A0A1W4WPX5 B4NJV5 A0A1A9VM15 A0A1B0AHF0 A0A1B0GDY2 B4GE28 D6W6W8 A0A182RA03 A0A182N5T6 A0A084WRG2 A0A182QTF8 N6UJA5 A0A182PAE0 B3M1Z1 B4KBG6 B3P6S4 F5HK36 A0A182XI20 A0A182HT03 A0A182XWX6 A0A182SFZ9 A0A182LVV2 A0A182VXD0 A0A2R7WXR3 A0A1J1HKM7 A0A182FT34 A0A182JBJ8 E0VH79 A0A182G9Q1 Q1DGX0
Pubmed
EMBL
BABH01007574
JTDY01000195
KOB78353.1
ODYU01000659
SOQ35819.1
KQ460226
+ More
KPJ16440.1 NWSH01002700 PCG67658.1 KZ150649 KZ149992 PZC70415.1 PZC75530.1 KQ459597 KPI95034.1 AGBW02010703 OWR48007.1 NWSH01000568 PCG75592.1 PCG75599.1 PZC75531.1 OWR48006.1 KPJ16441.1 ODYU01000495 SOQ35369.1 KPI95035.1 KOB78351.1 KPJ16439.1 KPI95033.1 DS232254 EDS38506.1 PCG67656.1 PCG67657.1 KZ149960 PZC76348.1 NEVH01007821 PNF35342.1 KK852563 KDR21300.1 JXUM01081455 KQ563248 KXJ74211.1 JXUM01028133 KQ560811 KXJ80795.1 AJVK01024455 CH477258 EAT45803.1 PYGN01000329 PSN48418.1 JXJN01020071 CH964272 EDW83957.1 CCAG010012073 CH479182 EDW33863.1 KQ971307 EFA11474.1 ATLV01026048 KE525405 KFB52806.1 AXCN02001155 APGK01022008 KB740396 KB631825 ENN80746.1 ERL86550.1 CH902617 EDV42251.1 CH933806 EDW13633.1 CH954182 EDV53744.1 AAAB01008859 EGK96877.1 APCN01000388 AXCM01005216 KK855810 PTY23960.1 CVRI01000008 CRK88483.1 DS235161 EEB12735.1 JXUM01049633 KQ561614 KXJ77997.1 CH900120 EAT32400.1 EAT45804.1
KPJ16440.1 NWSH01002700 PCG67658.1 KZ150649 KZ149992 PZC70415.1 PZC75530.1 KQ459597 KPI95034.1 AGBW02010703 OWR48007.1 NWSH01000568 PCG75592.1 PCG75599.1 PZC75531.1 OWR48006.1 KPJ16441.1 ODYU01000495 SOQ35369.1 KPI95035.1 KOB78351.1 KPJ16439.1 KPI95033.1 DS232254 EDS38506.1 PCG67656.1 PCG67657.1 KZ149960 PZC76348.1 NEVH01007821 PNF35342.1 KK852563 KDR21300.1 JXUM01081455 KQ563248 KXJ74211.1 JXUM01028133 KQ560811 KXJ80795.1 AJVK01024455 CH477258 EAT45803.1 PYGN01000329 PSN48418.1 JXJN01020071 CH964272 EDW83957.1 CCAG010012073 CH479182 EDW33863.1 KQ971307 EFA11474.1 ATLV01026048 KE525405 KFB52806.1 AXCN02001155 APGK01022008 KB740396 KB631825 ENN80746.1 ERL86550.1 CH902617 EDV42251.1 CH933806 EDW13633.1 CH954182 EDV53744.1 AAAB01008859 EGK96877.1 APCN01000388 AXCM01005216 KK855810 PTY23960.1 CVRI01000008 CRK88483.1 DS235161 EEB12735.1 JXUM01049633 KQ561614 KXJ77997.1 CH900120 EAT32400.1 EAT45804.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000218220
UP000053268
UP000007151
+ More
UP000002320 UP000235965 UP000027135 UP000069940 UP000249989 UP000075902 UP000092462 UP000008820 UP000075903 UP000245037 UP000091820 UP000092443 UP000092460 UP000075882 UP000192223 UP000007798 UP000078200 UP000092445 UP000092444 UP000008744 UP000007266 UP000075900 UP000075884 UP000030765 UP000075886 UP000019118 UP000030742 UP000075885 UP000007801 UP000009192 UP000008711 UP000007062 UP000076407 UP000075840 UP000076408 UP000075901 UP000075883 UP000075920 UP000183832 UP000069272 UP000075880 UP000009046
UP000002320 UP000235965 UP000027135 UP000069940 UP000249989 UP000075902 UP000092462 UP000008820 UP000075903 UP000245037 UP000091820 UP000092443 UP000092460 UP000075882 UP000192223 UP000007798 UP000078200 UP000092445 UP000092444 UP000008744 UP000007266 UP000075900 UP000075884 UP000030765 UP000075886 UP000019118 UP000030742 UP000075885 UP000007801 UP000009192 UP000008711 UP000007062 UP000076407 UP000075840 UP000076408 UP000075901 UP000075883 UP000075920 UP000183832 UP000069272 UP000075880 UP000009046
PRIDE
Pfam
PF00250 Forkhead
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
CDD
ProteinModelPortal
H9J210
A0A0L7LSA3
A0A2H1V4U3
A0A0N1IAY8
A0A2A4J7M3
A0A2W1B6E1
+ More
A0A194PQL6 A0A212F2P8 A0A2A4JW33 A0A2W1BML0 A0A212F2P9 A0A0N0PDE0 A0A2H1V3J5 A0A194PV77 A0A0L7LSZ6 A0A0N0PDS3 A0A194PPY8 B0X157 A0A2A4J7E0 A0A2W1BN29 A0A2J7R3D1 A0A067RL19 A0A182GQV0 A0A182H7J4 A0A182TKZ5 A0A1B0GME1 Q17GJ8 A0A182V4W8 A0A2P8YVZ1 A0A1A9W2Q1 A0A1A9Y7F8 A0A1B0BTB3 A0A182LF41 A0A1W4WPX5 B4NJV5 A0A1A9VM15 A0A1B0AHF0 A0A1B0GDY2 B4GE28 D6W6W8 A0A182RA03 A0A182N5T6 A0A084WRG2 A0A182QTF8 N6UJA5 A0A182PAE0 B3M1Z1 B4KBG6 B3P6S4 F5HK36 A0A182XI20 A0A182HT03 A0A182XWX6 A0A182SFZ9 A0A182LVV2 A0A182VXD0 A0A2R7WXR3 A0A1J1HKM7 A0A182FT34 A0A182JBJ8 E0VH79 A0A182G9Q1 Q1DGX0
A0A194PQL6 A0A212F2P8 A0A2A4JW33 A0A2W1BML0 A0A212F2P9 A0A0N0PDE0 A0A2H1V3J5 A0A194PV77 A0A0L7LSZ6 A0A0N0PDS3 A0A194PPY8 B0X157 A0A2A4J7E0 A0A2W1BN29 A0A2J7R3D1 A0A067RL19 A0A182GQV0 A0A182H7J4 A0A182TKZ5 A0A1B0GME1 Q17GJ8 A0A182V4W8 A0A2P8YVZ1 A0A1A9W2Q1 A0A1A9Y7F8 A0A1B0BTB3 A0A182LF41 A0A1W4WPX5 B4NJV5 A0A1A9VM15 A0A1B0AHF0 A0A1B0GDY2 B4GE28 D6W6W8 A0A182RA03 A0A182N5T6 A0A084WRG2 A0A182QTF8 N6UJA5 A0A182PAE0 B3M1Z1 B4KBG6 B3P6S4 F5HK36 A0A182XI20 A0A182HT03 A0A182XWX6 A0A182SFZ9 A0A182LVV2 A0A182VXD0 A0A2R7WXR3 A0A1J1HKM7 A0A182FT34 A0A182JBJ8 E0VH79 A0A182G9Q1 Q1DGX0
PDB
6FEC
E-value=1.03016e-34,
Score=365
Ontologies
KEGG
GO
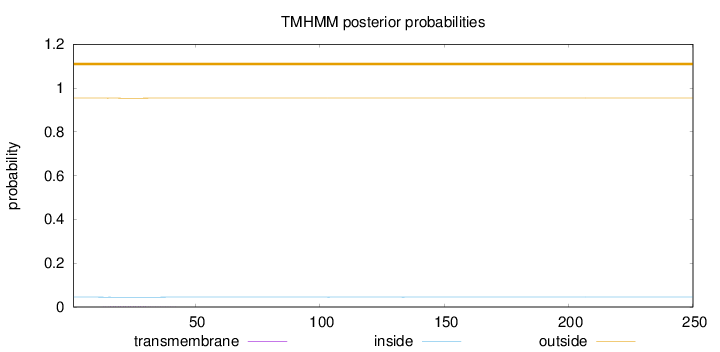
Topology
Subcellular location
Nucleus
Length:
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0342500000000001
Exp number, first 60 AAs:
0.03187
Total prob of N-in:
0.04538
outside
1 - 250
Population Genetic Test Statistics
Pi
266.755631
Theta
228.797508
Tajima's D
0.522465
CLR
0.157787
CSRT
0.518574071296435
Interpretation
Uncertain
