Gene
KWMTBOMO02914 Validated by peptides from experiments
Annotation
PREDICTED:_electron_transfer_flavoprotein_subunit_alpha?_mitochondrial_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.748
Sequence
CDS
ATGTTTGCACCCAATAGCAGGCATTTATTTTTGTCTGGACAGCTGCGTCGTTTGCAAAGTACTTTAGTGCTCGCCGAGCACAACAATGAGGTACTATCTCCAGCAACCCAAAATACATTGAATGCTGCCAAGAAAATTGGAGGTGATGTCTCAGTGCTTGTGGCTGGAACCAAATGTGGACCTGCTGCAGAATCCATTGCTAAGGCAAATGGTATATCTAAAGTTTTAGTGGCTGAGAGTGATGTCTTTAAAGGTTTTACTGCTGAAACCCTGACTCCTCTGATTTTGGCTACACAAAAACAGTTCAAATTCACTCATATCTTAGCACCAGCAACTGCTTTCGGAAAGACGGTGCTGCCACGAGTTGCGGCCAAGCTTGATGTGTCACCAATAACTGATATCATTGGTATCAAGGATGCTAATACCTTTGTGAGAACAATTTATGCTGGTAACGCTATATTAACCCTTGAGGCCAAAGATCCTATCAAGGTCATCACTGTTCGTGGCACAGCATTCCCTGCAGAGCCATTGGAGGGAGGGTCTGCAGCAATTGATAAAGCACCTGAGGGCGACTATAAGACTGATCTGGTTGAATTTTTAAAGCAGGAATTGACCAAATCTGACAGACCAGAACTCACTAGTGCTAAGAATATTGTATCTGGTGGACGAGGCTTGAAGTCTGGTGACAACTTCAAGTTGTTGTATGATCTAGCTGATAAGTTGAATGCGGCCGTGGGTGCCTCTCGTGCAGCTGTCGATGCTGGCTTTGTACCCAACGACCTACAGATTGGACAGACTGGCAAGATTGTTGCTCCTGACTTGTACATCGCTGTAGGCATCAGTGGTGCTATTCAGCACTTGGCCGGCATGAAAGACTCCAAAACCATTGTAGCAATCAACAAAGATCCTGAAGCTCCGATTTTCCAGGTGTCAGACTACGGTTTAGTCGCGGACTTGTTCAAGGCTGTTCCAGAACTGACTTCAAAGTTGTAA
Protein
MFAPNSRHLFLSGQLRRLQSTLVLAEHNNEVLSPATQNTLNAAKKIGGDVSVLVAGTKCGPAAESIAKANGISKVLVAESDVFKGFTAETLTPLILATQKQFKFTHILAPATAFGKTVLPRVAAKLDVSPITDIIGIKDANTFVRTIYAGNAILTLEAKDPIKVITVRGTAFPAEPLEGGSAAIDKAPEGDYKTDLVEFLKQELTKSDRPELTSAKNIVSGGRGLKSGDNFKLLYDLADKLNAAVGASRAAVDAGFVPNDLQIGQTGKIVAPDLYIAVGISGAIQHLAGMKDSKTIVAINKDPEAPIFQVSDYGLVADLFKAVPELTSKL
Summary
Uniprot
A0A194PNZ5
A0A2W1BMR8
A0A2A4J6E7
A0A2H1WJW3
A0A212F969
A0A0N0PDH1
+ More
A0A0K8TP49 A0A0M3QUI1 B4LIV9 A0A1Y1KBW9 D6W6F9 B3MCW4 A0A1L8EEC1 A0A1W4VBY4 A0A0L0BTK3 J3JYP8 A0A1I8ML35 B4N5R0 B4KUG0 T1PFC4 A0A0U2PGV9 B4JVR7 Q7KN94 Q7KLW5 W8AMN4 B3NSD7 A0A336LE55 B4P620 B4QBN6 A0A034VEG4 A0A0C9PGT3 A0A3B0JQM1 A0A0A1X9L9 Q28XD0 B4H5B6 A0A0K8TW28 A0A1I8PJC9 A0A023EPW1 Q2LIY7 A0A182G804 A0A088A872 A0A154P579 V5GSY6 D3TP14 A0A2J7PVT5 A0A2A3E8L2 B0W927 A0A1A9WQY6 A0A232EKZ2 A0A1W4X157 A0A1L8E0T6 A0A1L8E0V9 A0A067RBU8 A0A1A9XIE6 A0A1B0B1V9 A0A1A9V1T1 A0A1B6EG33 A0A1Q3FHQ1 A0A182KML5 A0A182UDW3 Q7Q254 A0A026W0B9 A0A182I867 A0A182XKH9 A0A182US61 A0A182NDX7 A0A182QT32 A0A182RSL2 A0A182VWZ8 Q16IA8 E0VFL6 A0A182IKL9 A0A0L7QVT2 E9IDX6 A0A182P432 A0A1B0G725 U5EIM4 A0A1I9WLC9 A0A195BWW2 A0A158NGX4 A0A182T7A2 E2ABK5 A0A182MVX3 A0A182YG09 F4WZH9 K7J9P1 W5JKP6 A0A195DRF2 A0A182FA34 A0A084VTK9 A0A1B6I6I5 A0A1B6F2A1 A0A151X297 A0A023EMX6 A0A2M4AAB1 A0A195CV73 A0A2M3Z3P5 A0A2M4BTR4 U4UL42 A0A1J1HVB3
A0A0K8TP49 A0A0M3QUI1 B4LIV9 A0A1Y1KBW9 D6W6F9 B3MCW4 A0A1L8EEC1 A0A1W4VBY4 A0A0L0BTK3 J3JYP8 A0A1I8ML35 B4N5R0 B4KUG0 T1PFC4 A0A0U2PGV9 B4JVR7 Q7KN94 Q7KLW5 W8AMN4 B3NSD7 A0A336LE55 B4P620 B4QBN6 A0A034VEG4 A0A0C9PGT3 A0A3B0JQM1 A0A0A1X9L9 Q28XD0 B4H5B6 A0A0K8TW28 A0A1I8PJC9 A0A023EPW1 Q2LIY7 A0A182G804 A0A088A872 A0A154P579 V5GSY6 D3TP14 A0A2J7PVT5 A0A2A3E8L2 B0W927 A0A1A9WQY6 A0A232EKZ2 A0A1W4X157 A0A1L8E0T6 A0A1L8E0V9 A0A067RBU8 A0A1A9XIE6 A0A1B0B1V9 A0A1A9V1T1 A0A1B6EG33 A0A1Q3FHQ1 A0A182KML5 A0A182UDW3 Q7Q254 A0A026W0B9 A0A182I867 A0A182XKH9 A0A182US61 A0A182NDX7 A0A182QT32 A0A182RSL2 A0A182VWZ8 Q16IA8 E0VFL6 A0A182IKL9 A0A0L7QVT2 E9IDX6 A0A182P432 A0A1B0G725 U5EIM4 A0A1I9WLC9 A0A195BWW2 A0A158NGX4 A0A182T7A2 E2ABK5 A0A182MVX3 A0A182YG09 F4WZH9 K7J9P1 W5JKP6 A0A195DRF2 A0A182FA34 A0A084VTK9 A0A1B6I6I5 A0A1B6F2A1 A0A151X297 A0A023EMX6 A0A2M4AAB1 A0A195CV73 A0A2M3Z3P5 A0A2M4BTR4 U4UL42 A0A1J1HVB3
Pubmed
26354079
28756777
22118469
26369729
17994087
28004739
+ More
18362917 19820115 18057021 26108605 22516182 23537049 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 17550304 22936249 25348373 25830018 15632085 24945155 26483478 20353571 28648823 24845553 20966253 12364791 14747013 17210077 24508170 30249741 17510324 20566863 21282665 27538518 21347285 20798317 25244985 21719571 20075255 20920257 23761445 24438588
18362917 19820115 18057021 26108605 22516182 23537049 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 17550304 22936249 25348373 25830018 15632085 24945155 26483478 20353571 28648823 24845553 20966253 12364791 14747013 17210077 24508170 30249741 17510324 20566863 21282665 27538518 21347285 20798317 25244985 21719571 20075255 20920257 23761445 24438588
EMBL
KQ459597
KPI95032.1
KZ149960
PZC76349.1
NWSH01002700
PCG67655.1
+ More
ODYU01009133 SOQ53339.1 AGBW02010703 AGBW02009627 OWR48009.1 OWR50284.1 KQ460226 KPJ16438.1 GDAI01001451 JAI16152.1 CP012524 ALC40709.1 CH940648 EDW60409.1 GEZM01086819 JAV58973.1 KQ971307 EFA11105.1 CH902619 EDV37366.1 KPU76755.1 GFDG01001737 JAV17062.1 JRES01001352 KNC23390.1 APGK01024802 BT128378 KB740562 AEE63336.1 ENN80305.1 CH964154 EDW79699.1 CH933808 EDW10747.1 KRG05455.1 KA647472 AFP62101.1 KR181956 ALJ76680.1 CH916375 EDV98055.1 AF132155 AE013599 AAD34743.1 AAF58627.1 AAM68717.1 AF160913 AAD46853.2 GAMC01016530 JAB90025.1 CH954179 EDV56439.1 UFQS01003574 UFQT01003574 SSX15769.1 SSX35120.1 CM000158 EDW90895.1 CM000362 CM002911 EDX06653.1 KMY93028.1 GAKP01018455 GAKP01018454 JAC40497.1 GBYB01000083 JAG69850.1 OUUW01000001 SPP74961.1 GBXI01006651 JAD07641.1 CM000071 ENO01916.1 CH479210 EDW32952.1 GDHF01033647 GDHF01010534 GDHF01009479 GDHF01005398 JAI18667.1 JAI41780.1 JAI42835.1 JAI46916.1 GAPW01002539 GAPW01002538 JAC11060.1 DQ256399 ABC61672.1 JXUM01047202 KQ561510 KXJ78340.1 KQ434820 KZC07013.1 GALX01001167 JAB67299.1 EZ423166 ADD19442.1 NEVH01020943 PNF20443.1 KZ288325 PBC28048.1 DS231861 EDS39465.1 NNAY01003676 OXU19015.1 GFDF01001746 JAV12338.1 GFDF01001747 JAV12337.1 KK852607 KDR20339.1 JXJN01007356 GEDC01000396 JAS36902.1 GFDL01007955 JAV27090.1 AAAB01008978 EAA13601.4 KK107519 QOIP01000009 EZA49370.1 RLU19121.1 APCN01002659 AXCN02002211 CH478091 EAT34001.1 DS235116 EEB12172.1 KQ414721 KOC62733.1 GL762556 EFZ21179.1 CCAG010003762 GANO01002575 JAB57296.1 KU932313 APA33949.1 KQ976398 KYM92775.1 ADTU01001619 GL438297 EFN69176.1 AXCM01002744 GL888479 EGI60423.1 AAZX01008083 ADMH02001262 ETN63349.1 KQ980612 KYN15084.1 ATLV01016361 KE525079 KFB41303.1 GECU01025172 GECU01013664 JAS82534.1 JAS94042.1 GECZ01025492 JAS44277.1 KQ982585 KYQ54376.1 GAPW01002881 JAC10717.1 GGFK01004388 MBW37709.1 KQ977259 KYN04530.1 GGFM01002386 MBW23137.1 GGFJ01007252 MBW56393.1 KB632399 ERL94794.1 CVRI01000021 CRK91490.1
ODYU01009133 SOQ53339.1 AGBW02010703 AGBW02009627 OWR48009.1 OWR50284.1 KQ460226 KPJ16438.1 GDAI01001451 JAI16152.1 CP012524 ALC40709.1 CH940648 EDW60409.1 GEZM01086819 JAV58973.1 KQ971307 EFA11105.1 CH902619 EDV37366.1 KPU76755.1 GFDG01001737 JAV17062.1 JRES01001352 KNC23390.1 APGK01024802 BT128378 KB740562 AEE63336.1 ENN80305.1 CH964154 EDW79699.1 CH933808 EDW10747.1 KRG05455.1 KA647472 AFP62101.1 KR181956 ALJ76680.1 CH916375 EDV98055.1 AF132155 AE013599 AAD34743.1 AAF58627.1 AAM68717.1 AF160913 AAD46853.2 GAMC01016530 JAB90025.1 CH954179 EDV56439.1 UFQS01003574 UFQT01003574 SSX15769.1 SSX35120.1 CM000158 EDW90895.1 CM000362 CM002911 EDX06653.1 KMY93028.1 GAKP01018455 GAKP01018454 JAC40497.1 GBYB01000083 JAG69850.1 OUUW01000001 SPP74961.1 GBXI01006651 JAD07641.1 CM000071 ENO01916.1 CH479210 EDW32952.1 GDHF01033647 GDHF01010534 GDHF01009479 GDHF01005398 JAI18667.1 JAI41780.1 JAI42835.1 JAI46916.1 GAPW01002539 GAPW01002538 JAC11060.1 DQ256399 ABC61672.1 JXUM01047202 KQ561510 KXJ78340.1 KQ434820 KZC07013.1 GALX01001167 JAB67299.1 EZ423166 ADD19442.1 NEVH01020943 PNF20443.1 KZ288325 PBC28048.1 DS231861 EDS39465.1 NNAY01003676 OXU19015.1 GFDF01001746 JAV12338.1 GFDF01001747 JAV12337.1 KK852607 KDR20339.1 JXJN01007356 GEDC01000396 JAS36902.1 GFDL01007955 JAV27090.1 AAAB01008978 EAA13601.4 KK107519 QOIP01000009 EZA49370.1 RLU19121.1 APCN01002659 AXCN02002211 CH478091 EAT34001.1 DS235116 EEB12172.1 KQ414721 KOC62733.1 GL762556 EFZ21179.1 CCAG010003762 GANO01002575 JAB57296.1 KU932313 APA33949.1 KQ976398 KYM92775.1 ADTU01001619 GL438297 EFN69176.1 AXCM01002744 GL888479 EGI60423.1 AAZX01008083 ADMH02001262 ETN63349.1 KQ980612 KYN15084.1 ATLV01016361 KE525079 KFB41303.1 GECU01025172 GECU01013664 JAS82534.1 JAS94042.1 GECZ01025492 JAS44277.1 KQ982585 KYQ54376.1 GAPW01002881 JAC10717.1 GGFK01004388 MBW37709.1 KQ977259 KYN04530.1 GGFM01002386 MBW23137.1 GGFJ01007252 MBW56393.1 KB632399 ERL94794.1 CVRI01000021 CRK91490.1
Proteomes
UP000053268
UP000218220
UP000007151
UP000053240
UP000092553
UP000008792
+ More
UP000007266 UP000007801 UP000192221 UP000037069 UP000019118 UP000095301 UP000007798 UP000009192 UP000001070 UP000000803 UP000008711 UP000002282 UP000000304 UP000268350 UP000001819 UP000008744 UP000095300 UP000069940 UP000249989 UP000005203 UP000076502 UP000235965 UP000242457 UP000002320 UP000091820 UP000215335 UP000192223 UP000027135 UP000092443 UP000092460 UP000078200 UP000075882 UP000075902 UP000007062 UP000053097 UP000279307 UP000075840 UP000076407 UP000075903 UP000075884 UP000075886 UP000075900 UP000075920 UP000008820 UP000009046 UP000075880 UP000053825 UP000075885 UP000092444 UP000078540 UP000005205 UP000075901 UP000000311 UP000075883 UP000076408 UP000007755 UP000002358 UP000000673 UP000078492 UP000069272 UP000030765 UP000075809 UP000078542 UP000030742 UP000183832
UP000007266 UP000007801 UP000192221 UP000037069 UP000019118 UP000095301 UP000007798 UP000009192 UP000001070 UP000000803 UP000008711 UP000002282 UP000000304 UP000268350 UP000001819 UP000008744 UP000095300 UP000069940 UP000249989 UP000005203 UP000076502 UP000235965 UP000242457 UP000002320 UP000091820 UP000215335 UP000192223 UP000027135 UP000092443 UP000092460 UP000078200 UP000075882 UP000075902 UP000007062 UP000053097 UP000279307 UP000075840 UP000076407 UP000075903 UP000075884 UP000075886 UP000075900 UP000075920 UP000008820 UP000009046 UP000075880 UP000053825 UP000075885 UP000092444 UP000078540 UP000005205 UP000075901 UP000000311 UP000075883 UP000076408 UP000007755 UP000002358 UP000000673 UP000078492 UP000069272 UP000030765 UP000075809 UP000078542 UP000030742 UP000183832
PRIDE
Pfam
Interpro
IPR033947
ETF_alpha_N
+ More
IPR018206 ETF_asu_C_CS
IPR014730 ETF_a/b_N
IPR014729 Rossmann-like_a/b/a_fold
IPR001308 ETF_a/FixB
IPR014731 ETF_asu_C
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR025258 Zf-RING_9
IPR037213 Run_dom_sf
IPR004012 Run_dom
IPR002155 Thiolase
IPR020613 Thiolase_CS
IPR020610 Thiolase_AS
IPR020616 Thiolase_N
IPR016039 Thiolase-like
IPR020615 Thiolase_acyl_enz_int_AS
IPR020617 Thiolase_C
IPR018206 ETF_asu_C_CS
IPR014730 ETF_a/b_N
IPR014729 Rossmann-like_a/b/a_fold
IPR001308 ETF_a/FixB
IPR014731 ETF_asu_C
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR025258 Zf-RING_9
IPR037213 Run_dom_sf
IPR004012 Run_dom
IPR002155 Thiolase
IPR020613 Thiolase_CS
IPR020610 Thiolase_AS
IPR020616 Thiolase_N
IPR016039 Thiolase-like
IPR020615 Thiolase_acyl_enz_int_AS
IPR020617 Thiolase_C
Gene 3D
ProteinModelPortal
A0A194PNZ5
A0A2W1BMR8
A0A2A4J6E7
A0A2H1WJW3
A0A212F969
A0A0N0PDH1
+ More
A0A0K8TP49 A0A0M3QUI1 B4LIV9 A0A1Y1KBW9 D6W6F9 B3MCW4 A0A1L8EEC1 A0A1W4VBY4 A0A0L0BTK3 J3JYP8 A0A1I8ML35 B4N5R0 B4KUG0 T1PFC4 A0A0U2PGV9 B4JVR7 Q7KN94 Q7KLW5 W8AMN4 B3NSD7 A0A336LE55 B4P620 B4QBN6 A0A034VEG4 A0A0C9PGT3 A0A3B0JQM1 A0A0A1X9L9 Q28XD0 B4H5B6 A0A0K8TW28 A0A1I8PJC9 A0A023EPW1 Q2LIY7 A0A182G804 A0A088A872 A0A154P579 V5GSY6 D3TP14 A0A2J7PVT5 A0A2A3E8L2 B0W927 A0A1A9WQY6 A0A232EKZ2 A0A1W4X157 A0A1L8E0T6 A0A1L8E0V9 A0A067RBU8 A0A1A9XIE6 A0A1B0B1V9 A0A1A9V1T1 A0A1B6EG33 A0A1Q3FHQ1 A0A182KML5 A0A182UDW3 Q7Q254 A0A026W0B9 A0A182I867 A0A182XKH9 A0A182US61 A0A182NDX7 A0A182QT32 A0A182RSL2 A0A182VWZ8 Q16IA8 E0VFL6 A0A182IKL9 A0A0L7QVT2 E9IDX6 A0A182P432 A0A1B0G725 U5EIM4 A0A1I9WLC9 A0A195BWW2 A0A158NGX4 A0A182T7A2 E2ABK5 A0A182MVX3 A0A182YG09 F4WZH9 K7J9P1 W5JKP6 A0A195DRF2 A0A182FA34 A0A084VTK9 A0A1B6I6I5 A0A1B6F2A1 A0A151X297 A0A023EMX6 A0A2M4AAB1 A0A195CV73 A0A2M3Z3P5 A0A2M4BTR4 U4UL42 A0A1J1HVB3
A0A0K8TP49 A0A0M3QUI1 B4LIV9 A0A1Y1KBW9 D6W6F9 B3MCW4 A0A1L8EEC1 A0A1W4VBY4 A0A0L0BTK3 J3JYP8 A0A1I8ML35 B4N5R0 B4KUG0 T1PFC4 A0A0U2PGV9 B4JVR7 Q7KN94 Q7KLW5 W8AMN4 B3NSD7 A0A336LE55 B4P620 B4QBN6 A0A034VEG4 A0A0C9PGT3 A0A3B0JQM1 A0A0A1X9L9 Q28XD0 B4H5B6 A0A0K8TW28 A0A1I8PJC9 A0A023EPW1 Q2LIY7 A0A182G804 A0A088A872 A0A154P579 V5GSY6 D3TP14 A0A2J7PVT5 A0A2A3E8L2 B0W927 A0A1A9WQY6 A0A232EKZ2 A0A1W4X157 A0A1L8E0T6 A0A1L8E0V9 A0A067RBU8 A0A1A9XIE6 A0A1B0B1V9 A0A1A9V1T1 A0A1B6EG33 A0A1Q3FHQ1 A0A182KML5 A0A182UDW3 Q7Q254 A0A026W0B9 A0A182I867 A0A182XKH9 A0A182US61 A0A182NDX7 A0A182QT32 A0A182RSL2 A0A182VWZ8 Q16IA8 E0VFL6 A0A182IKL9 A0A0L7QVT2 E9IDX6 A0A182P432 A0A1B0G725 U5EIM4 A0A1I9WLC9 A0A195BWW2 A0A158NGX4 A0A182T7A2 E2ABK5 A0A182MVX3 A0A182YG09 F4WZH9 K7J9P1 W5JKP6 A0A195DRF2 A0A182FA34 A0A084VTK9 A0A1B6I6I5 A0A1B6F2A1 A0A151X297 A0A023EMX6 A0A2M4AAB1 A0A195CV73 A0A2M3Z3P5 A0A2M4BTR4 U4UL42 A0A1J1HVB3
PDB
2A1U
E-value=2.4174e-109,
Score=1010
Ontologies
GO
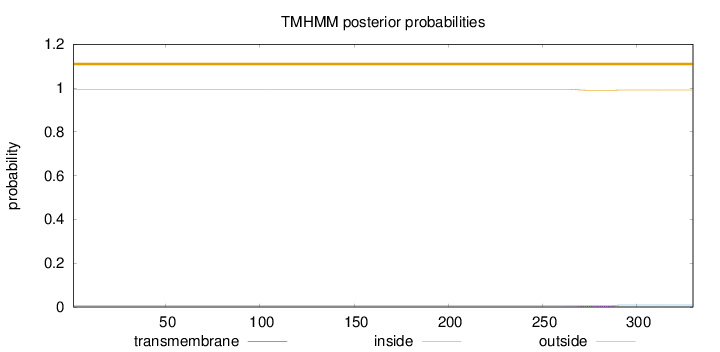
Topology
Length:
330
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08822
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.00564
outside
1 - 330
Population Genetic Test Statistics
Pi
238.678804
Theta
192.22864
Tajima's D
0.49107
CLR
0
CSRT
0.505824708764562
Interpretation
Uncertain
