Gene
KWMTBOMO02912
Pre Gene Modal
BGIBMGA003546
Annotation
hypothetical_protein_KGM_17655_[Danaus_plexippus]
Full name
Alkaline ceramidase
Alternative Name
Alkaline N-acylsphingosine amidohydrolase
Alkaline acylsphingosine deacylase
Protein brainwashing
Alkaline acylsphingosine deacylase
Protein brainwashing
Location in the cell
PlasmaMembrane Reliability : 4.999
Sequence
CDS
ATGTGGTCACATCTGGAACGTGGAAGCTCTCCGGTGGATTGGTGTGAATCAAATTATTCCATATCTCCTTTGATAGCAGAATTCGTTAACACTGTGAGCAATATTTTGTTCTTTCTGTTCCCACCAGTGTTGATTCATCTTTTCCAAGAATATTCAAAATTCTTCAATCCTGCTATCAATGTATTATGGGTTCTGTTGATGATTGTTGGACTTAGTTCAGCTTATTTTCATGCAACTTTGAGCTTAGTGGGTCAGTTATTAGATGAACTTGCAATATTGTGGGTATTTATGGCAGCATTTGCTATGTTCTTGCCTAAAAGATATTTTCCGAAATTTTTGGGAGGAAACAGGAGAATACTTGCACTGTACTCCAGTGTATTTTCGGTCATGTCGACTGGATTTCTCGTAATGCATCCAGCAGCTAATGCGTTCGCATTAATGACACTAGCTTTACCCGCCATGGGATTCTTGTGCAAGGAATTGACCAGAGTTAAATGCGCTCGTGTGTACCGTCTCGGGCTACGGTGTGTCGCTATCTGCTTCCTCGCAATGTTCTGTTGGATTATCGACCGTGTGTTTTGCGATGCTTGGCTCTCTATCAGCTTCCCTTACATGCACGGAATCTGGCATGTACTAATATTCATCGCCAGTTATACAGCACTCGTGCTTTTCGCGTATTTCAATGTCTCCGAGGAAAGGCCCGAACAAAAACCTCAATTGAAATATTGGCCGAGAAATGACTTTGAATTAGGCATACCTTATATAACCATAAAGCATCCTTGCAAAAAAGATTATAATATAGCAATATAG
Protein
MWSHLERGSSPVDWCESNYSISPLIAEFVNTVSNILFFLFPPVLIHLFQEYSKFFNPAINVLWVLLMIVGLSSAYFHATLSLVGQLLDELAILWVFMAAFAMFLPKRYFPKFLGGNRRILALYSSVFSVMSTGFLVMHPAANAFALMTLALPAMGFLCKELTRVKCARVYRLGLRCVAICFLAMFCWIIDRVFCDAWLSISFPYMHGIWHVLIFIASYTALVLFAYFNVSEERPEQKPQLKYWPRNDFELGIPYITIKHPCKKDYNIAI
Summary
Description
Hydrolyzes the sphingolipid ceramide into sphingosine and free fatty acid.
Catalytic Activity
an N-acylsphing-4-enine + H2O = a fatty acid + sphing-4-enine
Cofactor
Zn(2+)
Similarity
Belongs to the alkaline ceramidase family.
Keywords
Calcium
Complete proteome
Hydrolase
Lipid metabolism
Membrane
Metal-binding
Reference proteome
Transmembrane
Transmembrane helix
Zinc
Feature
chain Alkaline ceramidase
Uniprot
A0A212F961
A0A1E1WIZ4
A0A2A4J8D4
A0A2W1BQ00
A0A0N1IIJ1
H9J209
+ More
A0A067R9H6 A0A2J7PVS8 A0A310S639 A0A2A3ETE4 A0A088AJ78 E2BKT4 A0A232FN24 K7ITD4 A0A0L7RBV0 A0A154PLS3 A0A026WJ17 E9J4P4 A0A151HY84 A0A158NWA3 A0A195ET48 F4X4U0 A0A084VMJ7 A0A2M4BW77 A0A2M4AQ06 A0A182JGR3 A0A182F3S4 A0A2M3Z5F0 A0A2M4BWF2 A0A1Q3FNY5 A0A182U5B2 W5JTY4 A0A182K4W9 A0A182YLP2 A0A182KWH6 A0A182QHD4 A0A182R416 A0A182X684 Q7Q8A0 A0A182IBA5 A0A182NBB5 A0A182PVX1 B0W1A1 A0A182LWS7 Q17KM2 A0A182H840 A0A182VTH7 A0A1Y1LLY6 B3NL07 B4N7P1 B4PBH3 M9PDG7 A0A0J9R550 A0A1W4VK85 A0A1B0A1K4 D3TL44 I2CMG3 A0A1A9V9G9 A0A1A9XL29 A0A1B0B7Q0 B4I628 Q9VIP7 Q29KK7 B3ML29 A0A3B0JV50 A0A1I8PR19 A0A1A9W074 A0A336ML14 A0A336MLI1 B4KEP3 B4M9L6 B4JQ89 A0A0K8VIV9 A0A034VBN0 A0A0A1X4X2 A0A2P8YQ06 A0A151IC19 A0A151X5V1 A0A1I8MM63 E0VFL8 U5EWL1 A0A087SZP1 A0A1S3H206 A0A2L2YHP7 A0A2I9LNR0 A0A1D2N317 W5K4R2 M3ZUE7 A0A1W5AQM7 A0A0Q9X4L7 A0A0P9BNW0 A0A3B4E6G5 A0A1A7XQM8 A0A3B5L6N8 A0A2I4B6H7 A0A1A8ENS4 A0A1A8NBS1 A0A1A8JHJ8
A0A067R9H6 A0A2J7PVS8 A0A310S639 A0A2A3ETE4 A0A088AJ78 E2BKT4 A0A232FN24 K7ITD4 A0A0L7RBV0 A0A154PLS3 A0A026WJ17 E9J4P4 A0A151HY84 A0A158NWA3 A0A195ET48 F4X4U0 A0A084VMJ7 A0A2M4BW77 A0A2M4AQ06 A0A182JGR3 A0A182F3S4 A0A2M3Z5F0 A0A2M4BWF2 A0A1Q3FNY5 A0A182U5B2 W5JTY4 A0A182K4W9 A0A182YLP2 A0A182KWH6 A0A182QHD4 A0A182R416 A0A182X684 Q7Q8A0 A0A182IBA5 A0A182NBB5 A0A182PVX1 B0W1A1 A0A182LWS7 Q17KM2 A0A182H840 A0A182VTH7 A0A1Y1LLY6 B3NL07 B4N7P1 B4PBH3 M9PDG7 A0A0J9R550 A0A1W4VK85 A0A1B0A1K4 D3TL44 I2CMG3 A0A1A9V9G9 A0A1A9XL29 A0A1B0B7Q0 B4I628 Q9VIP7 Q29KK7 B3ML29 A0A3B0JV50 A0A1I8PR19 A0A1A9W074 A0A336ML14 A0A336MLI1 B4KEP3 B4M9L6 B4JQ89 A0A0K8VIV9 A0A034VBN0 A0A0A1X4X2 A0A2P8YQ06 A0A151IC19 A0A151X5V1 A0A1I8MM63 E0VFL8 U5EWL1 A0A087SZP1 A0A1S3H206 A0A2L2YHP7 A0A2I9LNR0 A0A1D2N317 W5K4R2 M3ZUE7 A0A1W5AQM7 A0A0Q9X4L7 A0A0P9BNW0 A0A3B4E6G5 A0A1A7XQM8 A0A3B5L6N8 A0A2I4B6H7 A0A1A8ENS4 A0A1A8NBS1 A0A1A8JHJ8
EC Number
3.5.1.-
3.5.1.23
3.5.1.23
Pubmed
22118469
28756777
26354079
19121390
24845553
20798317
+ More
28648823 20075255 24508170 30249741 21282665 21347285 21719571 24438588 20920257 23761445 25244985 20966253 12364791 14747013 17210077 17510324 26483478 28004739 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 20353571 22517621 12537569 10623899 14516700 15632085 25348373 25830018 29403074 25315136 20566863 26561354 29248469 27289101 25329095 23542700
28648823 20075255 24508170 30249741 21282665 21347285 21719571 24438588 20920257 23761445 25244985 20966253 12364791 14747013 17210077 17510324 26483478 28004739 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 20353571 22517621 12537569 10623899 14516700 15632085 25348373 25830018 29403074 25315136 20566863 26561354 29248469 27289101 25329095 23542700
EMBL
AGBW02009627
OWR50286.1
GDQN01004075
JAT86979.1
NWSH01002700
PCG67653.1
+ More
KZ149960 PZC76351.1 KQ460226 KPJ16436.1 BABH01007576 BABH01007577 BABH01007578 BABH01007579 KK852607 KDR20337.1 NEVH01020943 PNF20442.1 KQ768636 OAD53084.1 KZ288185 PBC34977.1 GL448826 EFN83683.1 NNAY01000006 OXU32131.1 AAZX01004114 KQ414617 KOC68231.1 KQ434977 KZC12831.1 KK107198 QOIP01000007 EZA55626.1 RLU21078.1 GL768112 EFZ12208.1 KQ976730 KYM76440.1 ADTU01027853 KQ981993 KYN31087.1 GL888679 EGI58513.1 ATLV01014599 KE524975 KFB39191.1 GGFJ01008194 MBW57335.1 GGFK01009555 MBW42876.1 GGFM01002991 MBW23742.1 GGFJ01008193 MBW57334.1 GFDL01005735 JAV29310.1 ADMH02000386 ETN66753.1 AXCN02000628 AAAB01008944 EAA10121.2 APCN01000661 DS231821 EDS44971.1 AXCM01000660 CH477223 EAT47204.1 JXUM01028611 KQ560826 KXJ80726.1 GEZM01051820 JAV74684.1 CH954179 EDV54585.2 CH964214 EDW80380.2 CM000158 EDW90487.2 AE014134 AGB93151.1 CM002910 KMY91121.1 EZ422142 ADD18339.1 CCAG010023642 JQ308541 AFJ68096.1 JXJN01009696 CH480822 EDW55834.1 AF323976 AY071232 CH379061 EAL33167.3 CH902620 EDV31647.2 OUUW01000010 SPP85975.1 UFQS01001128 UFQT01001128 SSX09146.1 SSX29057.1 UFQT01001296 SSX29879.1 CH933807 EDW11922.2 CH940654 EDW57892.2 CH916372 EDV99069.1 GDHF01013522 JAI38792.1 GAKP01019032 GAKP01019030 GAKP01019028 JAC39924.1 GBXI01008376 JAD05916.1 PYGN01000440 PSN46340.1 KQ978069 KYM97350.1 KQ982491 KYQ55756.1 DS235116 EEB12174.1 GANO01002907 JAB56964.1 KK112700 KFM58330.1 IAAA01021503 IAAA01021504 LAA07557.1 GFWZ01000033 MBW20023.1 LJIJ01000263 ODM99673.1 KRG02971.1 KPU73525.1 HADW01018981 SBP20381.1 HAEB01002288 SBQ48815.1 HAEH01001253 SBR66530.1 HAED01022679 HAEE01006814 SBR09443.1
KZ149960 PZC76351.1 KQ460226 KPJ16436.1 BABH01007576 BABH01007577 BABH01007578 BABH01007579 KK852607 KDR20337.1 NEVH01020943 PNF20442.1 KQ768636 OAD53084.1 KZ288185 PBC34977.1 GL448826 EFN83683.1 NNAY01000006 OXU32131.1 AAZX01004114 KQ414617 KOC68231.1 KQ434977 KZC12831.1 KK107198 QOIP01000007 EZA55626.1 RLU21078.1 GL768112 EFZ12208.1 KQ976730 KYM76440.1 ADTU01027853 KQ981993 KYN31087.1 GL888679 EGI58513.1 ATLV01014599 KE524975 KFB39191.1 GGFJ01008194 MBW57335.1 GGFK01009555 MBW42876.1 GGFM01002991 MBW23742.1 GGFJ01008193 MBW57334.1 GFDL01005735 JAV29310.1 ADMH02000386 ETN66753.1 AXCN02000628 AAAB01008944 EAA10121.2 APCN01000661 DS231821 EDS44971.1 AXCM01000660 CH477223 EAT47204.1 JXUM01028611 KQ560826 KXJ80726.1 GEZM01051820 JAV74684.1 CH954179 EDV54585.2 CH964214 EDW80380.2 CM000158 EDW90487.2 AE014134 AGB93151.1 CM002910 KMY91121.1 EZ422142 ADD18339.1 CCAG010023642 JQ308541 AFJ68096.1 JXJN01009696 CH480822 EDW55834.1 AF323976 AY071232 CH379061 EAL33167.3 CH902620 EDV31647.2 OUUW01000010 SPP85975.1 UFQS01001128 UFQT01001128 SSX09146.1 SSX29057.1 UFQT01001296 SSX29879.1 CH933807 EDW11922.2 CH940654 EDW57892.2 CH916372 EDV99069.1 GDHF01013522 JAI38792.1 GAKP01019032 GAKP01019030 GAKP01019028 JAC39924.1 GBXI01008376 JAD05916.1 PYGN01000440 PSN46340.1 KQ978069 KYM97350.1 KQ982491 KYQ55756.1 DS235116 EEB12174.1 GANO01002907 JAB56964.1 KK112700 KFM58330.1 IAAA01021503 IAAA01021504 LAA07557.1 GFWZ01000033 MBW20023.1 LJIJ01000263 ODM99673.1 KRG02971.1 KPU73525.1 HADW01018981 SBP20381.1 HAEB01002288 SBQ48815.1 HAEH01001253 SBR66530.1 HAED01022679 HAEE01006814 SBR09443.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000005204
UP000027135
UP000235965
+ More
UP000242457 UP000005203 UP000008237 UP000215335 UP000002358 UP000053825 UP000076502 UP000053097 UP000279307 UP000078540 UP000005205 UP000078541 UP000007755 UP000030765 UP000075880 UP000069272 UP000075902 UP000000673 UP000075881 UP000076408 UP000075882 UP000075886 UP000075900 UP000076407 UP000007062 UP000075840 UP000075884 UP000075885 UP000002320 UP000075883 UP000008820 UP000069940 UP000249989 UP000075920 UP000008711 UP000007798 UP000002282 UP000000803 UP000192221 UP000092445 UP000092444 UP000078200 UP000092443 UP000092460 UP000001292 UP000001819 UP000007801 UP000268350 UP000095300 UP000091820 UP000009192 UP000008792 UP000001070 UP000245037 UP000078542 UP000075809 UP000095301 UP000009046 UP000054359 UP000085678 UP000094527 UP000018467 UP000002852 UP000192224 UP000261440 UP000261380 UP000192220
UP000242457 UP000005203 UP000008237 UP000215335 UP000002358 UP000053825 UP000076502 UP000053097 UP000279307 UP000078540 UP000005205 UP000078541 UP000007755 UP000030765 UP000075880 UP000069272 UP000075902 UP000000673 UP000075881 UP000076408 UP000075882 UP000075886 UP000075900 UP000076407 UP000007062 UP000075840 UP000075884 UP000075885 UP000002320 UP000075883 UP000008820 UP000069940 UP000249989 UP000075920 UP000008711 UP000007798 UP000002282 UP000000803 UP000192221 UP000092445 UP000092444 UP000078200 UP000092443 UP000092460 UP000001292 UP000001819 UP000007801 UP000268350 UP000095300 UP000091820 UP000009192 UP000008792 UP000001070 UP000245037 UP000078542 UP000075809 UP000095301 UP000009046 UP000054359 UP000085678 UP000094527 UP000018467 UP000002852 UP000192224 UP000261440 UP000261380 UP000192220
Pfam
PF05875 Ceramidase
Interpro
IPR008901
ACER
ProteinModelPortal
A0A212F961
A0A1E1WIZ4
A0A2A4J8D4
A0A2W1BQ00
A0A0N1IIJ1
H9J209
+ More
A0A067R9H6 A0A2J7PVS8 A0A310S639 A0A2A3ETE4 A0A088AJ78 E2BKT4 A0A232FN24 K7ITD4 A0A0L7RBV0 A0A154PLS3 A0A026WJ17 E9J4P4 A0A151HY84 A0A158NWA3 A0A195ET48 F4X4U0 A0A084VMJ7 A0A2M4BW77 A0A2M4AQ06 A0A182JGR3 A0A182F3S4 A0A2M3Z5F0 A0A2M4BWF2 A0A1Q3FNY5 A0A182U5B2 W5JTY4 A0A182K4W9 A0A182YLP2 A0A182KWH6 A0A182QHD4 A0A182R416 A0A182X684 Q7Q8A0 A0A182IBA5 A0A182NBB5 A0A182PVX1 B0W1A1 A0A182LWS7 Q17KM2 A0A182H840 A0A182VTH7 A0A1Y1LLY6 B3NL07 B4N7P1 B4PBH3 M9PDG7 A0A0J9R550 A0A1W4VK85 A0A1B0A1K4 D3TL44 I2CMG3 A0A1A9V9G9 A0A1A9XL29 A0A1B0B7Q0 B4I628 Q9VIP7 Q29KK7 B3ML29 A0A3B0JV50 A0A1I8PR19 A0A1A9W074 A0A336ML14 A0A336MLI1 B4KEP3 B4M9L6 B4JQ89 A0A0K8VIV9 A0A034VBN0 A0A0A1X4X2 A0A2P8YQ06 A0A151IC19 A0A151X5V1 A0A1I8MM63 E0VFL8 U5EWL1 A0A087SZP1 A0A1S3H206 A0A2L2YHP7 A0A2I9LNR0 A0A1D2N317 W5K4R2 M3ZUE7 A0A1W5AQM7 A0A0Q9X4L7 A0A0P9BNW0 A0A3B4E6G5 A0A1A7XQM8 A0A3B5L6N8 A0A2I4B6H7 A0A1A8ENS4 A0A1A8NBS1 A0A1A8JHJ8
A0A067R9H6 A0A2J7PVS8 A0A310S639 A0A2A3ETE4 A0A088AJ78 E2BKT4 A0A232FN24 K7ITD4 A0A0L7RBV0 A0A154PLS3 A0A026WJ17 E9J4P4 A0A151HY84 A0A158NWA3 A0A195ET48 F4X4U0 A0A084VMJ7 A0A2M4BW77 A0A2M4AQ06 A0A182JGR3 A0A182F3S4 A0A2M3Z5F0 A0A2M4BWF2 A0A1Q3FNY5 A0A182U5B2 W5JTY4 A0A182K4W9 A0A182YLP2 A0A182KWH6 A0A182QHD4 A0A182R416 A0A182X684 Q7Q8A0 A0A182IBA5 A0A182NBB5 A0A182PVX1 B0W1A1 A0A182LWS7 Q17KM2 A0A182H840 A0A182VTH7 A0A1Y1LLY6 B3NL07 B4N7P1 B4PBH3 M9PDG7 A0A0J9R550 A0A1W4VK85 A0A1B0A1K4 D3TL44 I2CMG3 A0A1A9V9G9 A0A1A9XL29 A0A1B0B7Q0 B4I628 Q9VIP7 Q29KK7 B3ML29 A0A3B0JV50 A0A1I8PR19 A0A1A9W074 A0A336ML14 A0A336MLI1 B4KEP3 B4M9L6 B4JQ89 A0A0K8VIV9 A0A034VBN0 A0A0A1X4X2 A0A2P8YQ06 A0A151IC19 A0A151X5V1 A0A1I8MM63 E0VFL8 U5EWL1 A0A087SZP1 A0A1S3H206 A0A2L2YHP7 A0A2I9LNR0 A0A1D2N317 W5K4R2 M3ZUE7 A0A1W5AQM7 A0A0Q9X4L7 A0A0P9BNW0 A0A3B4E6G5 A0A1A7XQM8 A0A3B5L6N8 A0A2I4B6H7 A0A1A8ENS4 A0A1A8NBS1 A0A1A8JHJ8
PDB
6G7O
E-value=3.28877e-10,
Score=154
Ontologies
PATHWAY
GO
PANTHER
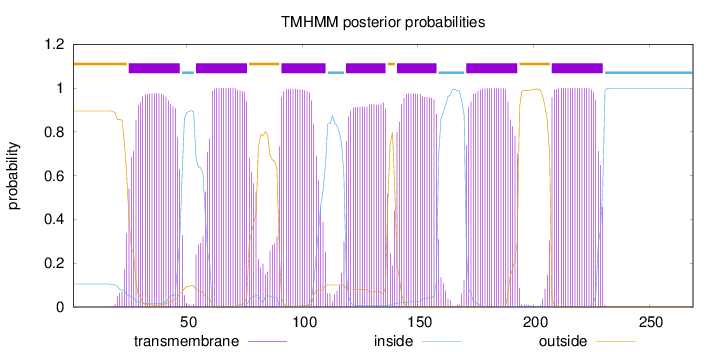
Topology
Subcellular location
Membrane
Length:
269
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
143.16278
Exp number, first 60 AAs:
24.711
Total prob of N-in:
0.10452
POSSIBLE N-term signal
sequence
outside
1 - 24
TMhelix
25 - 47
inside
48 - 53
TMhelix
54 - 76
outside
77 - 90
TMhelix
91 - 110
inside
111 - 118
TMhelix
119 - 136
outside
137 - 140
TMhelix
141 - 158
inside
159 - 170
TMhelix
171 - 193
outside
194 - 207
TMhelix
208 - 230
inside
231 - 269
Population Genetic Test Statistics
Pi
288.625308
Theta
169.083658
Tajima's D
2.51096
CLR
0.457631
CSRT
0.947102644867757
Interpretation
Uncertain
