Gene
KWMTBOMO02909
Annotation
PREDICTED:_proteasomal_ATPase-associated_factor_1-like_isoform_X2_[Plutella_xylostella]
Full name
Proteasomal ATPase-associated factor 1
Alternative Name
WD repeat-containing protein 71
Protein G-16
Protein G-16
Location in the cell
Extracellular Reliability : 1.118 PlasmaMembrane Reliability : 1.59
Sequence
CDS
ATGTCTCTACCTGATGCGAAGCCCGTCATCACCATCCAAAGTGATTGGAACTCTATTCTTCGGTTGCCCAATGGCCAGGTTTGGATATCCTCTAAAGTTTTAAACGAACTCAGTACACAAGAGACGTTAACAAGATCAGGAAACAAAAGTGATGGAAATATTAAGATAAATATTTCTAAAAATTATGAATACATTTCTCTTTCTAACTTAAGCCTTGTCGTGAAGCACATTGAATCTGAATTGAAAGCAGTGTTTGTTGCCCCCACCAAGGACCATAATATACATGATAAAGCAATTTTGTCTGTGGCACTTGCTGAAAACTCCTTAACTGTGTCTTCGTGTGAAGCAGATAAACTTCTTGTCTGGGATAGCAGGACAGGCGATCAGTTATTAGATTTAAAAGGACATGGTGGCCCAGTCTACAAATGCAGGTTCTTTCCGTCCGGAATTGTAGTACTGTCGGCAGGAGCAGATGGCTCATGTAGAATATGGTCAGCTGAGACAGGAATCAATCCTGTATTGTTAAAAGGACACACTATGTCAATCTCTGATATCAGTATCATTGACAGAGGGAATAATATTGTGTCTGTTAGCAGGGATGGTTCTGCAAAGCTTTGGGATGTCGGTAAATCCAATTGCTTAGATAATATTGTAGAGGGTCATGGACAAATAAATTGTTGCACAATTACAACAACAACAGACGAAGTTAAAGTTGATAACGAAAGAGAAGTTGGAACTTCAAATAAGCTATTAATTTTGGGCTTTGAAAGTGGTTTAATTATGTGTGCACATATAGCCAAACGACAAGAGTTGTTCACTAAGCAGATTGATTCAGCGTGTAATGCATGCATTGTCGTTGGTCAGTCTATTATTGTCGGTTGTAATAATGGCAAGGTTGTACAGTTGAATTTACAAGATGGAAATTTGATCAGTGAGTTATATGAATCCGCAAGTCCAGTTTTAAGTTTAACAATATTGGCAAATCAAATGTTCGCAATAGGACGCCAAGACGGAACGTGTACAGTGATATCATTACTAGAACAACACAAGAATACTAGGGTGTACCTAACTGGTCCAGACTGTGATGGTGTCAGAGATATAGCATTTAATGGAAAATGGATACTCACGGCGTGTCGAGATACAATAATCAGAAAATATGACTACAATCAAATCAACGTTCACTTTAAATGA
Protein
MSLPDAKPVITIQSDWNSILRLPNGQVWISSKVLNELSTQETLTRSGNKSDGNIKINISKNYEYISLSNLSLVVKHIESELKAVFVAPTKDHNIHDKAILSVALAENSLTVSSCEADKLLVWDSRTGDQLLDLKGHGGPVYKCRFFPSGIVVLSAGADGSCRIWSAETGINPVLLKGHTMSISDISIIDRGNNIVSVSRDGSAKLWDVGKSNCLDNIVEGHGQINCCTITTTTDEVKVDNEREVGTSNKLLILGFESGLIMCAHIAKRQELFTKQIDSACNACIVVGQSIIVGCNNGKVVQLNLQDGNLISELYESASPVLSLTILANQMFAIGRQDGTCTVISLLEQHKNTRVYLTGPDCDGVRDIAFNGKWILTACRDTIIRKYDYNQINVHFK
Summary
Description
Inhibits proteasome 26S assembly and activity by impairing the association of the 19S regulatory complex with the 20S core.
Inhibits proteasome 26S assembly and activity by impairing the association of the 19S regulatory complex with the 20S core. Protects SUPT6H from proteasomal degradation (By similarity).
Inhibits proteasome 26S assembly and proteolytic activity by impairing the association of the 19S regulatory complex with the 20S core. In case of HIV-1 infection, recruited by viral Tat to the HIV-1 promoter, where it promotes the recruitment of 19S regulatory complex through dissociation of the proteasome 26S. This presumably promotes provirus transcription efficiency. Protects SUPT6H from proteasomal degradation.
Inhibits proteasome 26S assembly and activity by impairing the association of the 19S regulatory complex with the 20S core. Protects SUPT6H from proteasomal degradation (By similarity).
Inhibits proteasome 26S assembly and proteolytic activity by impairing the association of the 19S regulatory complex with the 20S core. In case of HIV-1 infection, recruited by viral Tat to the HIV-1 promoter, where it promotes the recruitment of 19S regulatory complex through dissociation of the proteasome 26S. This presumably promotes provirus transcription efficiency. Protects SUPT6H from proteasomal degradation.
Subunit
Interacts with proteasome 26S subunit ATPases.
Interacts with PSMC1, PSMC2, PSMC3, PSMC4, PSMC5 and PSMC6. Interacts with SUPT6H.
(Microbial infection) Interacts with HIV-1 Tat.
Interacts with PSMC1, PSMC2, PSMC3, PSMC4, PSMC5 and PSMC6. Interacts with SUPT6H.
(Microbial infection) Interacts with HIV-1 Tat.
Similarity
Belongs to the WD repeat PAAF1/RPN14 family.
Keywords
Complete proteome
Proteasome
Reference proteome
Repeat
WD repeat
Acetylation
Alternative splicing
Host-virus interaction
Polymorphism
Feature
chain Proteasomal ATPase-associated factor 1
splice variant In isoform 3.
sequence variant In dbSNP:rs17850051.
splice variant In isoform 3.
sequence variant In dbSNP:rs17850051.
Uniprot
H9J208
A0A1E1W003
S4PX76
A0A1E1W2Q6
A0A2A4K1X2
A0A2W1BX75
+ More
A0A194PPY3 A0A2H1VPA0 A0A212F987 A0A0N1PIK9 A0A2J7RD98 A0A067RU69 A0A131XSB8 L7MJG5 A0A131YLL4 A0A1E1XSM4 B7P1K6 A0A224Z6J9 A0A2P8Y1D1 G3MHB7 A0A1B6C573 A0A0V0G5V1 E0VK37 A0A1B6ICC3 A0A1B6FQI2 R4G4J5 T1HFQ0 A0A1B6KVJ0 T1IYX6 A0A226E222 X1YV63 C3YX31 A0A1D2MTW9 A0A0P4VJK9 A0A023GJD7 W5MGF0 A0A2C9KV69 A0A224XTG1 A0A210QDW4 V4ADP7 A0A1S3HFZ7 H9GBE8 A0A146KWQ7 F1NPD7 A0A091GE79 Q5ZK69 A0A2I0M2N0 G1NQW2 A0A146M2V5 H2XTW5 A0A0A9WM83 A0A0K8SXE0 A0A0A0AY68 A0A2Y9DVD7 A0A218V328 K9J0H1 A0A093GQX2 G3WLC3 A0A2I2UZY0 G3ST40 F6TIQ4 Q6NUB1 A0A2K5F9S4 Q148I1 Q1JP70 Q9BRP4 Q5M7K8 A0A151PJR8 A0A2K5I4U3 A0A2Y9TGV1 A0A1W2W9U3 F7A0V2 I7GK76 A0A1L8F9L6 G7PN76 A0A0D9QVM5 A0A1U7U293 W5PEK0 A0A341CV56 A0A2Y9PRM6 A0A2S2QMD2 A0A2K5Z007 A0A2K6RLF7 A0A2K5RHM2 A0A3P4NG24 G9KEY6 A0A2U4C9L8 A0A2U3VBF2 A7RPH0 U6CS62 J9K557 A0A2Y9I8L5 A0A383ZFA6 G1T3M0 G1PJS8 G1MFW8 U3K024 A0A369RRT6 T2M702
A0A194PPY3 A0A2H1VPA0 A0A212F987 A0A0N1PIK9 A0A2J7RD98 A0A067RU69 A0A131XSB8 L7MJG5 A0A131YLL4 A0A1E1XSM4 B7P1K6 A0A224Z6J9 A0A2P8Y1D1 G3MHB7 A0A1B6C573 A0A0V0G5V1 E0VK37 A0A1B6ICC3 A0A1B6FQI2 R4G4J5 T1HFQ0 A0A1B6KVJ0 T1IYX6 A0A226E222 X1YV63 C3YX31 A0A1D2MTW9 A0A0P4VJK9 A0A023GJD7 W5MGF0 A0A2C9KV69 A0A224XTG1 A0A210QDW4 V4ADP7 A0A1S3HFZ7 H9GBE8 A0A146KWQ7 F1NPD7 A0A091GE79 Q5ZK69 A0A2I0M2N0 G1NQW2 A0A146M2V5 H2XTW5 A0A0A9WM83 A0A0K8SXE0 A0A0A0AY68 A0A2Y9DVD7 A0A218V328 K9J0H1 A0A093GQX2 G3WLC3 A0A2I2UZY0 G3ST40 F6TIQ4 Q6NUB1 A0A2K5F9S4 Q148I1 Q1JP70 Q9BRP4 Q5M7K8 A0A151PJR8 A0A2K5I4U3 A0A2Y9TGV1 A0A1W2W9U3 F7A0V2 I7GK76 A0A1L8F9L6 G7PN76 A0A0D9QVM5 A0A1U7U293 W5PEK0 A0A341CV56 A0A2Y9PRM6 A0A2S2QMD2 A0A2K5Z007 A0A2K6RLF7 A0A2K5RHM2 A0A3P4NG24 G9KEY6 A0A2U4C9L8 A0A2U3VBF2 A7RPH0 U6CS62 J9K557 A0A2Y9I8L5 A0A383ZFA6 G1T3M0 G1PJS8 G1MFW8 U3K024 A0A369RRT6 T2M702
Pubmed
19121390
23622113
28756777
26354079
22118469
24845553
+ More
25576852 26830274 29209593 28797301 29403074 22216098 20566863 23254933 18563158 27289101 27129103 15562597 28812685 21881562 26823975 15592404 15642098 23371554 20838655 12481130 15114417 25401762 21709235 17975172 20431018 11282978 16305752 14702039 16554811 15489334 15831487 17323924 17289585 19413330 21269460 22316138 22293439 17431167 22002653 25319552 17194215 27762356 20809919 25362486 23236062 17615350 21993624 20010809 30042472 24065732
25576852 26830274 29209593 28797301 29403074 22216098 20566863 23254933 18563158 27289101 27129103 15562597 28812685 21881562 26823975 15592404 15642098 23371554 20838655 12481130 15114417 25401762 21709235 17975172 20431018 11282978 16305752 14702039 16554811 15489334 15831487 17323924 17289585 19413330 21269460 22316138 22293439 17431167 22002653 25319552 17194215 27762356 20809919 25362486 23236062 17615350 21993624 20010809 30042472 24065732
EMBL
BABH01007580
BABH01007581
GDQN01010795
JAT80259.1
GAIX01005523
JAA87037.1
+ More
GDQN01009820 JAT81234.1 NWSH01000285 PCG77763.1 KZ149960 PZC76353.1 KQ459597 KPI95028.1 ODYU01003384 SOQ42074.1 AGBW02009627 OWR50288.1 KQ460226 KPJ16434.1 NEVH01005293 PNF38802.1 KK852463 KDR23389.1 GEFM01006836 JAP68960.1 GACK01001606 JAA63428.1 GEDV01009756 JAP78801.1 GFAA01001137 JAU02298.1 ABJB010162390 ABJB011131290 DS616928 EEC00478.1 GFPF01010814 MAA21960.1 PYGN01001052 PSN38067.1 JO841268 AEO32885.1 GEDC01028859 GEDC01025370 GEDC01022682 GEDC01018912 JAS08439.1 JAS11928.1 JAS14616.1 JAS18386.1 GECL01002665 JAP03459.1 DS235239 EEB13743.1 GECU01023125 JAS84581.1 GECZ01017302 JAS52467.1 GAHY01001305 JAA76205.1 ACPB03006513 ACPB03006514 ACPB03006515 GEBQ01024495 JAT15482.1 JH431701 LNIX01000009 OXA50526.1 AMQN01000169 GG666562 EEN55049.1 LJIJ01000571 ODM96145.1 GDKW01002206 JAI54389.1 GBBM01001394 JAC34024.1 AHAT01003069 GFTR01005167 JAW11259.1 NEDP02004067 OWF46858.1 KB201931 ESO93250.1 AAWZ02034903 GDHC01017708 JAQ00921.1 AADN05001070 KL448103 KFO80258.1 AJ720215 AKCR02000045 PKK23937.1 GDHC01004536 JAQ14093.1 EAAA01002310 GBHO01034057 GBHO01034056 JAG09547.1 JAG09548.1 GBRD01007858 JAG57963.1 KL873257 KGL98060.1 MUZQ01000065 OWK60131.1 GABZ01006501 JAA47024.1 KL216342 KFV69184.1 AEFK01119961 AEFK01119962 AEFK01119963 AEFK01119964 AEFK01119965 AANG04002890 AAMC01053271 AAMC01053272 AAMC01053273 BC068685 AAH68685.1 BC118307 BT025484 ABF57440.1 AF087895 AK021910 AK222501 AK298258 AK316394 AP002770 CH471076 BC006142 BC021541 BC028628 BC088584 AAH88584.1 AKHW03000111 KYO49084.1 JSUE03012924 JSUE03012925 JSUE03012926 JSUE03012927 JSUE03012928 JU322691 JU473233 CM001266 AFE66447.1 AFH30037.1 EHH23235.1 AB171255 BAE88318.1 CM004480 OCT68285.1 AQIA01021395 AQIA01021396 AQIA01021397 CM001289 EHH56571.1 AQIB01134136 AQIB01134137 AQIB01134138 AMGL01032426 AMGL01032427 AMGL01032428 GGMS01009645 MBY78848.1 CYRY02020271 VCW97011.1 JP014863 AES03461.1 DS469525 EDO46684.1 HAAF01000582 CCP72408.1 ABLF02039226 AAGW02008191 AAGW02008192 AAPE02040817 ACTA01002017 ACTA01186006 ACTA01194004 AGTO01011545 NOWV01000245 RDD37407.1 HAAD01001495 CDG67727.1
GDQN01009820 JAT81234.1 NWSH01000285 PCG77763.1 KZ149960 PZC76353.1 KQ459597 KPI95028.1 ODYU01003384 SOQ42074.1 AGBW02009627 OWR50288.1 KQ460226 KPJ16434.1 NEVH01005293 PNF38802.1 KK852463 KDR23389.1 GEFM01006836 JAP68960.1 GACK01001606 JAA63428.1 GEDV01009756 JAP78801.1 GFAA01001137 JAU02298.1 ABJB010162390 ABJB011131290 DS616928 EEC00478.1 GFPF01010814 MAA21960.1 PYGN01001052 PSN38067.1 JO841268 AEO32885.1 GEDC01028859 GEDC01025370 GEDC01022682 GEDC01018912 JAS08439.1 JAS11928.1 JAS14616.1 JAS18386.1 GECL01002665 JAP03459.1 DS235239 EEB13743.1 GECU01023125 JAS84581.1 GECZ01017302 JAS52467.1 GAHY01001305 JAA76205.1 ACPB03006513 ACPB03006514 ACPB03006515 GEBQ01024495 JAT15482.1 JH431701 LNIX01000009 OXA50526.1 AMQN01000169 GG666562 EEN55049.1 LJIJ01000571 ODM96145.1 GDKW01002206 JAI54389.1 GBBM01001394 JAC34024.1 AHAT01003069 GFTR01005167 JAW11259.1 NEDP02004067 OWF46858.1 KB201931 ESO93250.1 AAWZ02034903 GDHC01017708 JAQ00921.1 AADN05001070 KL448103 KFO80258.1 AJ720215 AKCR02000045 PKK23937.1 GDHC01004536 JAQ14093.1 EAAA01002310 GBHO01034057 GBHO01034056 JAG09547.1 JAG09548.1 GBRD01007858 JAG57963.1 KL873257 KGL98060.1 MUZQ01000065 OWK60131.1 GABZ01006501 JAA47024.1 KL216342 KFV69184.1 AEFK01119961 AEFK01119962 AEFK01119963 AEFK01119964 AEFK01119965 AANG04002890 AAMC01053271 AAMC01053272 AAMC01053273 BC068685 AAH68685.1 BC118307 BT025484 ABF57440.1 AF087895 AK021910 AK222501 AK298258 AK316394 AP002770 CH471076 BC006142 BC021541 BC028628 BC088584 AAH88584.1 AKHW03000111 KYO49084.1 JSUE03012924 JSUE03012925 JSUE03012926 JSUE03012927 JSUE03012928 JU322691 JU473233 CM001266 AFE66447.1 AFH30037.1 EHH23235.1 AB171255 BAE88318.1 CM004480 OCT68285.1 AQIA01021395 AQIA01021396 AQIA01021397 CM001289 EHH56571.1 AQIB01134136 AQIB01134137 AQIB01134138 AMGL01032426 AMGL01032427 AMGL01032428 GGMS01009645 MBY78848.1 CYRY02020271 VCW97011.1 JP014863 AES03461.1 DS469525 EDO46684.1 HAAF01000582 CCP72408.1 ABLF02039226 AAGW02008191 AAGW02008192 AAPE02040817 ACTA01002017 ACTA01186006 ACTA01194004 AGTO01011545 NOWV01000245 RDD37407.1 HAAD01001495 CDG67727.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000235965
+ More
UP000027135 UP000001555 UP000245037 UP000009046 UP000015103 UP000198287 UP000001554 UP000094527 UP000018468 UP000076420 UP000242188 UP000030746 UP000085678 UP000001646 UP000000539 UP000053760 UP000053872 UP000001645 UP000008144 UP000053858 UP000248480 UP000197619 UP000053875 UP000007648 UP000011712 UP000007646 UP000008143 UP000233020 UP000009136 UP000005640 UP000050525 UP000233080 UP000248484 UP000006718 UP000186698 UP000009130 UP000233100 UP000029965 UP000189704 UP000002356 UP000252040 UP000248483 UP000233140 UP000233200 UP000233040 UP000245320 UP000245340 UP000001593 UP000007819 UP000248481 UP000261681 UP000001811 UP000001074 UP000008912 UP000016665 UP000253843
UP000027135 UP000001555 UP000245037 UP000009046 UP000015103 UP000198287 UP000001554 UP000094527 UP000018468 UP000076420 UP000242188 UP000030746 UP000085678 UP000001646 UP000000539 UP000053760 UP000053872 UP000001645 UP000008144 UP000053858 UP000248480 UP000197619 UP000053875 UP000007648 UP000011712 UP000007646 UP000008143 UP000233020 UP000009136 UP000005640 UP000050525 UP000233080 UP000248484 UP000006718 UP000186698 UP000009130 UP000233100 UP000029965 UP000189704 UP000002356 UP000252040 UP000248483 UP000233140 UP000233200 UP000233040 UP000245320 UP000245340 UP000001593 UP000007819 UP000248481 UP000261681 UP000001811 UP000001074 UP000008912 UP000016665 UP000253843
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9J208
A0A1E1W003
S4PX76
A0A1E1W2Q6
A0A2A4K1X2
A0A2W1BX75
+ More
A0A194PPY3 A0A2H1VPA0 A0A212F987 A0A0N1PIK9 A0A2J7RD98 A0A067RU69 A0A131XSB8 L7MJG5 A0A131YLL4 A0A1E1XSM4 B7P1K6 A0A224Z6J9 A0A2P8Y1D1 G3MHB7 A0A1B6C573 A0A0V0G5V1 E0VK37 A0A1B6ICC3 A0A1B6FQI2 R4G4J5 T1HFQ0 A0A1B6KVJ0 T1IYX6 A0A226E222 X1YV63 C3YX31 A0A1D2MTW9 A0A0P4VJK9 A0A023GJD7 W5MGF0 A0A2C9KV69 A0A224XTG1 A0A210QDW4 V4ADP7 A0A1S3HFZ7 H9GBE8 A0A146KWQ7 F1NPD7 A0A091GE79 Q5ZK69 A0A2I0M2N0 G1NQW2 A0A146M2V5 H2XTW5 A0A0A9WM83 A0A0K8SXE0 A0A0A0AY68 A0A2Y9DVD7 A0A218V328 K9J0H1 A0A093GQX2 G3WLC3 A0A2I2UZY0 G3ST40 F6TIQ4 Q6NUB1 A0A2K5F9S4 Q148I1 Q1JP70 Q9BRP4 Q5M7K8 A0A151PJR8 A0A2K5I4U3 A0A2Y9TGV1 A0A1W2W9U3 F7A0V2 I7GK76 A0A1L8F9L6 G7PN76 A0A0D9QVM5 A0A1U7U293 W5PEK0 A0A341CV56 A0A2Y9PRM6 A0A2S2QMD2 A0A2K5Z007 A0A2K6RLF7 A0A2K5RHM2 A0A3P4NG24 G9KEY6 A0A2U4C9L8 A0A2U3VBF2 A7RPH0 U6CS62 J9K557 A0A2Y9I8L5 A0A383ZFA6 G1T3M0 G1PJS8 G1MFW8 U3K024 A0A369RRT6 T2M702
A0A194PPY3 A0A2H1VPA0 A0A212F987 A0A0N1PIK9 A0A2J7RD98 A0A067RU69 A0A131XSB8 L7MJG5 A0A131YLL4 A0A1E1XSM4 B7P1K6 A0A224Z6J9 A0A2P8Y1D1 G3MHB7 A0A1B6C573 A0A0V0G5V1 E0VK37 A0A1B6ICC3 A0A1B6FQI2 R4G4J5 T1HFQ0 A0A1B6KVJ0 T1IYX6 A0A226E222 X1YV63 C3YX31 A0A1D2MTW9 A0A0P4VJK9 A0A023GJD7 W5MGF0 A0A2C9KV69 A0A224XTG1 A0A210QDW4 V4ADP7 A0A1S3HFZ7 H9GBE8 A0A146KWQ7 F1NPD7 A0A091GE79 Q5ZK69 A0A2I0M2N0 G1NQW2 A0A146M2V5 H2XTW5 A0A0A9WM83 A0A0K8SXE0 A0A0A0AY68 A0A2Y9DVD7 A0A218V328 K9J0H1 A0A093GQX2 G3WLC3 A0A2I2UZY0 G3ST40 F6TIQ4 Q6NUB1 A0A2K5F9S4 Q148I1 Q1JP70 Q9BRP4 Q5M7K8 A0A151PJR8 A0A2K5I4U3 A0A2Y9TGV1 A0A1W2W9U3 F7A0V2 I7GK76 A0A1L8F9L6 G7PN76 A0A0D9QVM5 A0A1U7U293 W5PEK0 A0A341CV56 A0A2Y9PRM6 A0A2S2QMD2 A0A2K5Z007 A0A2K6RLF7 A0A2K5RHM2 A0A3P4NG24 G9KEY6 A0A2U4C9L8 A0A2U3VBF2 A7RPH0 U6CS62 J9K557 A0A2Y9I8L5 A0A383ZFA6 G1T3M0 G1PJS8 G1MFW8 U3K024 A0A369RRT6 T2M702
PDB
3VL1
E-value=6.06192e-19,
Score=231
Ontologies
GO
Topology
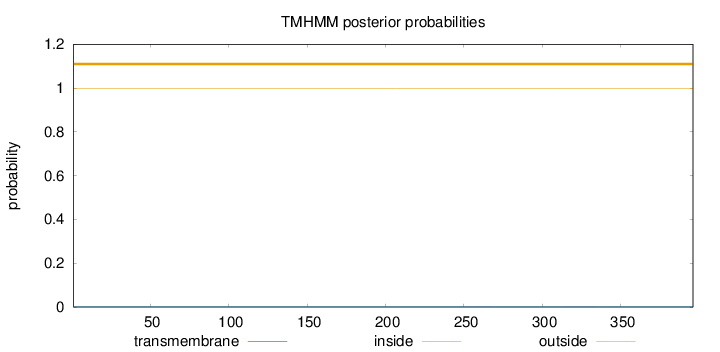
Length:
396
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01315
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00314
outside
1 - 396
Population Genetic Test Statistics
Pi
215.150436
Theta
147.181237
Tajima's D
1.137958
CLR
0.271765
CSRT
0.697915104244788
Interpretation
Uncertain
