Gene
KWMTBOMO02908
Pre Gene Modal
BGIBMGA003644
Annotation
PREDICTED:_sorting_nexin-17_isoform_X1_[Amyelois_transitella]
Full name
Sorting nexin-17
Location in the cell
Nuclear Reliability : 3.783
Sequence
CDS
ATGCATTTCTCAATACCTGATTTACAACAATTTTACGATGACAATGGTACTTCATATACGGGATACAATATATATATTGATGGATTTTTCCATTGCACCGCCCGCTATAGACAGTTGCTCAGTCTCCATGAGCAGCTGCAAGCACAACACTCATATGTTAGACTTCCACAATTTCCACCGAAGAAGTTGTTCCTTACCAGTTCACTGCTGGAAGAAAGGAGGATCCTGTTAGAGAAATATATACAAATGGTGGGTCAAATTCCTGTGCTGGCTAATTCAGATTTACTTATAACATTCCTGTTTTCTGCTCAACAAGAAACTCACTCTGTTAAAATGCATGAAGTTGAAATAGAAATATCCTTAATGAATGGCTATAGAATACCATTGTCTGTTTCTTCCACTGATTCATCAAGTACTTTACTGGATATTGCTTGTAATTACATTAATTTATCAAGTGAATTAATAAAATATTATTCATTATATTTATTTAACTGGAGTTGTGCTAAAGATGGACAACCTTCAATGAAAAAATTAGAAGAATATGAATCTCCCTACATATCACAAAAATATGTGAGACCCGAAGATAAAATAGTTTTAAGAAAAAGCTACTGGGATCCTAATTATGACGTTGATCTTATGATTGACAATGTATCTTTAGACTTGTTATATCTACAAACAAATGAAGAGCTAGATCTTGGCTGGATAATTGCAGATGCTCAAACTAGAGACATTTTAAGAAAATATGAAGCTAAAAAACAAAAAAGAGAGTTTATGGAATTAGCGCGCTCGTTGAGACATTACGGTCGTATTCCAGCTGGAGAAGCGGTGACAGACTCAGTAAACGTATCTGGAGATAATTCTGACAGCACGTGTCGTGTTCGTGTTTCTCTTGCTGCCAGGGAACTGACTCTGACCACGGTCACTCAGCCTCAAAGGGAACAACGGTTTAAAGTTACACGCATGAGATGTTGGAGAATCACGACGTTGCACTGCATGGAAAGGCCACAAACTAACGGACATAGTTCAGAAAACGAAGAACCAAATAAAAACTTTGAATTATCCTTTGAATATCTAATTAGCAAGGATAATTTACAGTGGATAACCTTGAGGACGGAACATGCCACATTCATAAGCGTCTGTTTACAGTCAATAGTTGACGAGTTGATGCGGCAGAAAAGCGGCGCAGGACCAACGTCCCCTCGATGCGAGAAGCGCACCCATCTCACATATTTGAGGAGGGACGGCTCCAGTCAACTTATCTCGCCGTCCTCCTCCAGCGATACGCTCAGTTCTAACAATGCCGACTCTTCATACAACTCAACTAGTTCCAAGGAAATATTTTCAGTTCAAAGACTAACCGAGAAATTCGCAACTGTCGCCTTCAAAACCGGAAAGGACTGTGTTGAAAATAACGCTTTCGAAGCTATCGGCGATGAGGAATTGTAA
Protein
MHFSIPDLQQFYDDNGTSYTGYNIYIDGFFHCTARYRQLLSLHEQLQAQHSYVRLPQFPPKKLFLTSSLLEERRILLEKYIQMVGQIPVLANSDLLITFLFSAQQETHSVKMHEVEIEISLMNGYRIPLSVSSTDSSSTLLDIACNYINLSSELIKYYSLYLFNWSCAKDGQPSMKKLEEYESPYISQKYVRPEDKIVLRKSYWDPNYDVDLMIDNVSLDLLYLQTNEELDLGWIIADAQTRDILRKYEAKKQKREFMELARSLRHYGRIPAGEAVTDSVNVSGDNSDSTCRVRVSLAARELTLTTVTQPQREQRFKVTRMRCWRITTLHCMERPQTNGHSSENEEPNKNFELSFEYLISKDNLQWITLRTEHATFISVCLQSIVDELMRQKSGAGPTSPRCEKRTHLTYLRRDGSSQLISPSSSSDTLSSNNADSSYNSTSSKEIFSVQRLTEKFATVAFKTGKDCVENNAFEAIGDEEL
Summary
Description
Critical regulator of endosomal recycling of numerous surface proteins, including integrins, signaling receptor and channels. Binds to NPxY sequences in the cytoplasmic tails of target cargos. Associates with retriever and CCC complexes to prevent lysosomal degradation and promote cell surface recycling of numerous cargos such as integrins ITGB1, ITGB5 and their associated alpha subunits. Also required for maintenance of normal cell surface levels of APP and LRP1. Interacts with membranes containing phosphatidylinositol 3-phosphate (PtdIns(3P)).
Critical regulator of endosomal recycling of numerous surface proteins, including integrins, signaling receptor and channels (PubMed:15121882, PubMed:15769472). Binds to NPxY sequences in the cytoplasmic tails of target cargos (PubMed:21512128). Associates with retriever and CCC complexes to prevent lysosomal degradation and promote cell surface recycling of numerous cargos such as integrins ITGB1, ITGB5 and their associated alpha subunits (PubMed:28892079, PubMed:22492727). Also required for maintenance of normal cell surface levels of APP and LRP1 (PubMed:16712798, PubMed:19005208). Interacts with membranes containing phosphatidylinositol 3-phosphate (PtdIns(3P)) (PubMed:16712798).
Critical regulator of endosomal recycling of numerous surface proteins, including integrins, signaling receptor and channels (PubMed:15121882, PubMed:15769472). Binds to NPxY sequences in the cytoplasmic tails of target cargos (PubMed:21512128). Associates with retriever and CCC complexes to prevent lysosomal degradation and promote cell surface recycling of numerous cargos such as integrins ITGB1, ITGB5 and their associated alpha subunits (PubMed:28892079, PubMed:22492727). Also required for maintenance of normal cell surface levels of APP and LRP1 (PubMed:16712798, PubMed:19005208). Interacts with membranes containing phosphatidylinositol 3-phosphate (PtdIns(3P)) (PubMed:16712798).
Subunit
Monomer (By similarity). Interacts with CCDC22, CCDC93, DSCR3 and VPS35L; the interaction with DSCR3 is direct and associates SNX17 with the retriever and CCC complexes (By similarity).
Monomer (By similarity). Interacts with APP (via cytoplasmic YXNPXY motif). Interacts with KIF1B (By similarity). Interacts with the C-termini of P-selectin, PTC, LDLR, VLDLR, LRP1 and LRP8. Interacts with KRIT1 (via N-terminus). Interacts with HRAS. Interacts with ITGB1 and ITGB5 (via NPxY motif). Interacts with CCDC22, CCDC93, VPS26C and VPS35L; the interaction with VPS26C is direct and associates SNX17 with the retriever and CCC complexes (By similarity).
Monomer (PubMed:21512128). Interacts with APP (via cytoplasmic YXNPXY motif) (By similarity). Interacts with KIF1B (By similarity). Interacts with the C-termini of P-selectin, PTC, LDLR, VLDLR, LRP1 and LRP8 (PubMed:11237770, PubMed:14739284, PubMed:15769472, PubMed:19005208). Interacts with KRIT1 (via N-terminus) (PubMed:16712798). Interacts with HRAS (PubMed:21512128). Interacts with ITGB1 and ITGB5 (via NPxY motif) (PubMed:22492727). Interacts with CCDC22, CCDC93, VPS26C and VPS35L; the interaction with VPS26C is direct and associates SNX17 with the retriever and CCC complexes (PubMed:28892079).
Monomer (By similarity). Interacts with APP (via cytoplasmic YXNPXY motif). Interacts with KIF1B (By similarity). Interacts with the C-termini of P-selectin, PTC, LDLR, VLDLR, LRP1 and LRP8. Interacts with KRIT1 (via N-terminus). Interacts with HRAS. Interacts with ITGB1 and ITGB5 (via NPxY motif). Interacts with CCDC22, CCDC93, VPS26C and VPS35L; the interaction with VPS26C is direct and associates SNX17 with the retriever and CCC complexes (By similarity).
Monomer (PubMed:21512128). Interacts with APP (via cytoplasmic YXNPXY motif) (By similarity). Interacts with KIF1B (By similarity). Interacts with the C-termini of P-selectin, PTC, LDLR, VLDLR, LRP1 and LRP8 (PubMed:11237770, PubMed:14739284, PubMed:15769472, PubMed:19005208). Interacts with KRIT1 (via N-terminus) (PubMed:16712798). Interacts with HRAS (PubMed:21512128). Interacts with ITGB1 and ITGB5 (via NPxY motif) (PubMed:22492727). Interacts with CCDC22, CCDC93, VPS26C and VPS35L; the interaction with VPS26C is direct and associates SNX17 with the retriever and CCC complexes (PubMed:28892079).
Similarity
Belongs to the sorting nexin family.
Keywords
Complete proteome
Cytoplasm
Cytoplasmic vesicle
Endosome
Lipid-binding
Membrane
Protein transport
Reference proteome
Transport
Phosphoprotein
3D-structure
Alternative splicing
Direct protein sequencing
Feature
chain Sorting nexin-17
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J2A7
A0A2A4K1J0
A0A2W1BRT3
A0A2H1VMM6
A0A194PNX5
A0A0N1PIS0
+ More
A0A212F9A2 A0A0T6B469 A0A1Y1LRH0 A0A1Y1LTE6 A0A067RCT0 A0A2J7QKV5 A0A1L8DW83 A0A1B6GLU6 A0A0P4VUB6 A0A0K8TSY8 A0A139WNV0 U5EWH9 N6TXU9 T1HCC1 A0A069DUD8 A0A0V0G6X8 A0A023EX41 A0A182JDB1 A0A1Q3FM90 A0A1Q3FM63 E0W474 W5JF46 A0A2M4BLC7 A0A1Q3FM11 A0A084WB98 A0A1Q3FMA9 Q174K7 A0A224XND6 A0A1Q3FM12 A0A182HNH2 A0A1Q3FLS6 A0A182LPD6 A0A182XC71 A0A182R8M1 A0A1B6M1P5 A0A182WF87 A0A182MY18 A0A182URN9 A0A182PVD1 A0A0A9WCV1 Q7Q353 A0A182MUF3 A0A336ML37 A0A182YK00 A0A182FU18 A0A1J1HEI5 K7J1R5 A0A232FDG1 Q5RID7 A0A0R4ITX6 A0A091CUU5 A0A1S3JSU6 A0A182U5R5 G1QTR3 A0A1S3EZ36 F7EIH8 A0A2K6EJR1 G5BZ07 H0X6Q1 A0A336MZ61 A0A250YIQ9 L5KRI9 G1TCZ5 A0A384AVI1 A0A2F0BBG3 I3MH81 A0A2Y9ETK1 A0A1S3I4B5 K9ISV1 E2RFE5 Q5EA77 A0A1S3AN44 G1NVG7 A0A337SPJ6 A0A2Y9KQN9 A0A3P4S0S0 M3YED2 A0A2K5YKU2 A0A2Y9HMR7 A0A2U3Z2H9 G1L3Y3 H0VAS9 A0A2R5LC33 A0A2K5D0S8 A0A2K6ANR0 A0A2K5X0R5 H2P6N6 Q15036 F6XSD1 A0A182KFR3 F7IBA9
A0A212F9A2 A0A0T6B469 A0A1Y1LRH0 A0A1Y1LTE6 A0A067RCT0 A0A2J7QKV5 A0A1L8DW83 A0A1B6GLU6 A0A0P4VUB6 A0A0K8TSY8 A0A139WNV0 U5EWH9 N6TXU9 T1HCC1 A0A069DUD8 A0A0V0G6X8 A0A023EX41 A0A182JDB1 A0A1Q3FM90 A0A1Q3FM63 E0W474 W5JF46 A0A2M4BLC7 A0A1Q3FM11 A0A084WB98 A0A1Q3FMA9 Q174K7 A0A224XND6 A0A1Q3FM12 A0A182HNH2 A0A1Q3FLS6 A0A182LPD6 A0A182XC71 A0A182R8M1 A0A1B6M1P5 A0A182WF87 A0A182MY18 A0A182URN9 A0A182PVD1 A0A0A9WCV1 Q7Q353 A0A182MUF3 A0A336ML37 A0A182YK00 A0A182FU18 A0A1J1HEI5 K7J1R5 A0A232FDG1 Q5RID7 A0A0R4ITX6 A0A091CUU5 A0A1S3JSU6 A0A182U5R5 G1QTR3 A0A1S3EZ36 F7EIH8 A0A2K6EJR1 G5BZ07 H0X6Q1 A0A336MZ61 A0A250YIQ9 L5KRI9 G1TCZ5 A0A384AVI1 A0A2F0BBG3 I3MH81 A0A2Y9ETK1 A0A1S3I4B5 K9ISV1 E2RFE5 Q5EA77 A0A1S3AN44 G1NVG7 A0A337SPJ6 A0A2Y9KQN9 A0A3P4S0S0 M3YED2 A0A2K5YKU2 A0A2Y9HMR7 A0A2U3Z2H9 G1L3Y3 H0VAS9 A0A2R5LC33 A0A2K5D0S8 A0A2K6ANR0 A0A2K5X0R5 H2P6N6 Q15036 F6XSD1 A0A182KFR3 F7IBA9
Pubmed
19121390
28756777
26354079
22118469
28004739
24845553
+ More
27129103 26369729 18362917 19820115 23537049 26334808 25474469 20566863 20920257 23761445 24438588 17510324 20966253 25401762 26823975 12364791 25244985 20075255 28648823 23594743 17495919 21993625 28087693 23258410 21993624 16341006 16305752 17975172 20010809 7584044 14702039 15815621 15489334 12665801 11237770 14739284 15121882 15769472 16712798 18669648 19005208 19690332 20068231 21269460 21406692 22492727 23186163 24275569 28892079 21512128 19892987 25243066
27129103 26369729 18362917 19820115 23537049 26334808 25474469 20566863 20920257 23761445 24438588 17510324 20966253 25401762 26823975 12364791 25244985 20075255 28648823 23594743 17495919 21993625 28087693 23258410 21993624 16341006 16305752 17975172 20010809 7584044 14702039 15815621 15489334 12665801 11237770 14739284 15121882 15769472 16712798 18669648 19005208 19690332 20068231 21269460 21406692 22492727 23186163 24275569 28892079 21512128 19892987 25243066
EMBL
BABH01007581
BABH01007582
BABH01007583
NWSH01000285
PCG77764.1
KZ149960
+ More
PZC76354.1 ODYU01003384 SOQ42073.1 KQ459597 KPI95012.1 KQ460226 KPJ16420.1 AGBW02009627 OWR50289.1 LJIG01015994 KRT81934.1 GEZM01050507 JAV75571.1 GEZM01050508 JAV75570.1 KK852549 KDR21557.1 NEVH01013256 PNF29213.1 GFDF01003392 JAV10692.1 GECZ01006350 JAS63419.1 GDKW01000324 JAI56271.1 GDAI01000350 JAI17253.1 KQ971307 KYB29604.1 GANO01000563 JAB59308.1 APGK01047095 KB741077 KB632384 ENN74125.1 ERL94256.1 ACPB03009391 GBGD01001319 JAC87570.1 GECL01002249 JAP03875.1 GBBI01004904 JAC13808.1 GFDL01006316 JAV28729.1 GFDL01006469 JAV28576.1 DS235886 EEB20430.1 ADMH02001328 ETN62992.1 GGFJ01004726 MBW53867.1 GFDL01006512 JAV28533.1 ATLV01022328 KE525331 KFB47492.1 GFDL01006295 JAV28750.1 CH477409 EAT41496.1 GFTR01006334 JAW10092.1 GFDL01006519 JAV28526.1 APCN01001930 GFDL01006571 JAV28474.1 GEBQ01010116 JAT29861.1 GBHO01038388 GBHO01038387 GBHO01038386 GBRD01017635 GDHC01017179 JAG05216.1 JAG05217.1 JAG05218.1 JAG48192.1 JAQ01450.1 AAAB01008966 EAA12955.4 AXCM01000360 UFQT01001603 SSX31134.1 CVRI01000001 CRK86367.1 NNAY01000427 OXU28479.1 BX276101 BC162993 KN123775 KFO23339.1 ADFV01061229 ADFV01061230 JH172504 GEBF01003470 EHB14518.1 JAO00163.1 AAQR03049365 UFQS01004128 UFQT01004128 SSX16194.1 SSX35520.1 GFFW01001317 JAV43471.1 KB030580 ELK14072.1 AAGW02005694 NTJE010126768 MBW00349.1 AGTP01025101 GABZ01001502 JAA52023.1 AAEX03010804 BT020692 BC118249 AAPE02039359 AANG04003329 CYRY02044488 VCX39458.1 AEYP01063354 ACTA01192627 AAKN02023460 GGLE01002781 MBY06907.1 AQIA01018733 ABGA01389649 NDHI03003415 PNJ58928.1 D31764 AJ404855 AJ404856 BT007167 AK298869 CR457081 AK222543 AC074117 BC002524 BC002610 BC014620 BC050590 GAMT01008022 GAMS01008786 GAMR01003658 GAMQ01000214 GAMP01000658 JAB03839.1 JAB14350.1 JAB30274.1 JAB41637.1 JAB52097.1
PZC76354.1 ODYU01003384 SOQ42073.1 KQ459597 KPI95012.1 KQ460226 KPJ16420.1 AGBW02009627 OWR50289.1 LJIG01015994 KRT81934.1 GEZM01050507 JAV75571.1 GEZM01050508 JAV75570.1 KK852549 KDR21557.1 NEVH01013256 PNF29213.1 GFDF01003392 JAV10692.1 GECZ01006350 JAS63419.1 GDKW01000324 JAI56271.1 GDAI01000350 JAI17253.1 KQ971307 KYB29604.1 GANO01000563 JAB59308.1 APGK01047095 KB741077 KB632384 ENN74125.1 ERL94256.1 ACPB03009391 GBGD01001319 JAC87570.1 GECL01002249 JAP03875.1 GBBI01004904 JAC13808.1 GFDL01006316 JAV28729.1 GFDL01006469 JAV28576.1 DS235886 EEB20430.1 ADMH02001328 ETN62992.1 GGFJ01004726 MBW53867.1 GFDL01006512 JAV28533.1 ATLV01022328 KE525331 KFB47492.1 GFDL01006295 JAV28750.1 CH477409 EAT41496.1 GFTR01006334 JAW10092.1 GFDL01006519 JAV28526.1 APCN01001930 GFDL01006571 JAV28474.1 GEBQ01010116 JAT29861.1 GBHO01038388 GBHO01038387 GBHO01038386 GBRD01017635 GDHC01017179 JAG05216.1 JAG05217.1 JAG05218.1 JAG48192.1 JAQ01450.1 AAAB01008966 EAA12955.4 AXCM01000360 UFQT01001603 SSX31134.1 CVRI01000001 CRK86367.1 NNAY01000427 OXU28479.1 BX276101 BC162993 KN123775 KFO23339.1 ADFV01061229 ADFV01061230 JH172504 GEBF01003470 EHB14518.1 JAO00163.1 AAQR03049365 UFQS01004128 UFQT01004128 SSX16194.1 SSX35520.1 GFFW01001317 JAV43471.1 KB030580 ELK14072.1 AAGW02005694 NTJE010126768 MBW00349.1 AGTP01025101 GABZ01001502 JAA52023.1 AAEX03010804 BT020692 BC118249 AAPE02039359 AANG04003329 CYRY02044488 VCX39458.1 AEYP01063354 ACTA01192627 AAKN02023460 GGLE01002781 MBY06907.1 AQIA01018733 ABGA01389649 NDHI03003415 PNJ58928.1 D31764 AJ404855 AJ404856 BT007167 AK298869 CR457081 AK222543 AC074117 BC002524 BC002610 BC014620 BC050590 GAMT01008022 GAMS01008786 GAMR01003658 GAMQ01000214 GAMP01000658 JAB03839.1 JAB14350.1 JAB30274.1 JAB41637.1 JAB52097.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000027135
+ More
UP000235965 UP000007266 UP000019118 UP000030742 UP000015103 UP000075880 UP000009046 UP000000673 UP000030765 UP000008820 UP000075840 UP000075882 UP000076407 UP000075900 UP000075920 UP000075884 UP000075903 UP000075885 UP000007062 UP000075883 UP000076408 UP000069272 UP000183832 UP000002358 UP000215335 UP000000437 UP000028990 UP000085678 UP000075902 UP000001073 UP000081671 UP000002280 UP000233160 UP000006813 UP000005225 UP000010552 UP000001811 UP000261681 UP000005215 UP000248484 UP000002254 UP000009136 UP000079721 UP000001074 UP000011712 UP000248482 UP000000715 UP000233140 UP000248481 UP000245341 UP000008912 UP000005447 UP000233020 UP000233120 UP000233100 UP000001595 UP000005640 UP000002281 UP000075881 UP000008225
UP000235965 UP000007266 UP000019118 UP000030742 UP000015103 UP000075880 UP000009046 UP000000673 UP000030765 UP000008820 UP000075840 UP000075882 UP000076407 UP000075900 UP000075920 UP000075884 UP000075903 UP000075885 UP000007062 UP000075883 UP000076408 UP000069272 UP000183832 UP000002358 UP000215335 UP000000437 UP000028990 UP000085678 UP000075902 UP000001073 UP000081671 UP000002280 UP000233160 UP000006813 UP000005225 UP000010552 UP000001811 UP000261681 UP000005215 UP000248484 UP000002254 UP000009136 UP000079721 UP000001074 UP000011712 UP000248482 UP000000715 UP000233140 UP000248481 UP000245341 UP000008912 UP000005447 UP000233020 UP000233120 UP000233100 UP000001595 UP000005640 UP000002281 UP000075881 UP000008225
Interpro
IPR037831
SNX17/27/31
+ More
IPR037836 SNX17_FERM-like_dom
IPR011993 PH-like_dom_sf
IPR040842 SNX17/31_FERM
IPR001683 Phox
IPR036871 PX_dom_sf
IPR028666 SNX17
IPR035963 FERM_2
IPR019749 Band_41_domain
IPR000159 RA_dom
IPR019748 FERM_central
IPR000299 FERM_domain
IPR020846 MFS_dom
IPR039672 MFS_2
IPR036259 MFS_trans_sf
IPR037836 SNX17_FERM-like_dom
IPR011993 PH-like_dom_sf
IPR040842 SNX17/31_FERM
IPR001683 Phox
IPR036871 PX_dom_sf
IPR028666 SNX17
IPR035963 FERM_2
IPR019749 Band_41_domain
IPR000159 RA_dom
IPR019748 FERM_central
IPR000299 FERM_domain
IPR020846 MFS_dom
IPR039672 MFS_2
IPR036259 MFS_trans_sf
Gene 3D
ProteinModelPortal
H9J2A7
A0A2A4K1J0
A0A2W1BRT3
A0A2H1VMM6
A0A194PNX5
A0A0N1PIS0
+ More
A0A212F9A2 A0A0T6B469 A0A1Y1LRH0 A0A1Y1LTE6 A0A067RCT0 A0A2J7QKV5 A0A1L8DW83 A0A1B6GLU6 A0A0P4VUB6 A0A0K8TSY8 A0A139WNV0 U5EWH9 N6TXU9 T1HCC1 A0A069DUD8 A0A0V0G6X8 A0A023EX41 A0A182JDB1 A0A1Q3FM90 A0A1Q3FM63 E0W474 W5JF46 A0A2M4BLC7 A0A1Q3FM11 A0A084WB98 A0A1Q3FMA9 Q174K7 A0A224XND6 A0A1Q3FM12 A0A182HNH2 A0A1Q3FLS6 A0A182LPD6 A0A182XC71 A0A182R8M1 A0A1B6M1P5 A0A182WF87 A0A182MY18 A0A182URN9 A0A182PVD1 A0A0A9WCV1 Q7Q353 A0A182MUF3 A0A336ML37 A0A182YK00 A0A182FU18 A0A1J1HEI5 K7J1R5 A0A232FDG1 Q5RID7 A0A0R4ITX6 A0A091CUU5 A0A1S3JSU6 A0A182U5R5 G1QTR3 A0A1S3EZ36 F7EIH8 A0A2K6EJR1 G5BZ07 H0X6Q1 A0A336MZ61 A0A250YIQ9 L5KRI9 G1TCZ5 A0A384AVI1 A0A2F0BBG3 I3MH81 A0A2Y9ETK1 A0A1S3I4B5 K9ISV1 E2RFE5 Q5EA77 A0A1S3AN44 G1NVG7 A0A337SPJ6 A0A2Y9KQN9 A0A3P4S0S0 M3YED2 A0A2K5YKU2 A0A2Y9HMR7 A0A2U3Z2H9 G1L3Y3 H0VAS9 A0A2R5LC33 A0A2K5D0S8 A0A2K6ANR0 A0A2K5X0R5 H2P6N6 Q15036 F6XSD1 A0A182KFR3 F7IBA9
A0A212F9A2 A0A0T6B469 A0A1Y1LRH0 A0A1Y1LTE6 A0A067RCT0 A0A2J7QKV5 A0A1L8DW83 A0A1B6GLU6 A0A0P4VUB6 A0A0K8TSY8 A0A139WNV0 U5EWH9 N6TXU9 T1HCC1 A0A069DUD8 A0A0V0G6X8 A0A023EX41 A0A182JDB1 A0A1Q3FM90 A0A1Q3FM63 E0W474 W5JF46 A0A2M4BLC7 A0A1Q3FM11 A0A084WB98 A0A1Q3FMA9 Q174K7 A0A224XND6 A0A1Q3FM12 A0A182HNH2 A0A1Q3FLS6 A0A182LPD6 A0A182XC71 A0A182R8M1 A0A1B6M1P5 A0A182WF87 A0A182MY18 A0A182URN9 A0A182PVD1 A0A0A9WCV1 Q7Q353 A0A182MUF3 A0A336ML37 A0A182YK00 A0A182FU18 A0A1J1HEI5 K7J1R5 A0A232FDG1 Q5RID7 A0A0R4ITX6 A0A091CUU5 A0A1S3JSU6 A0A182U5R5 G1QTR3 A0A1S3EZ36 F7EIH8 A0A2K6EJR1 G5BZ07 H0X6Q1 A0A336MZ61 A0A250YIQ9 L5KRI9 G1TCZ5 A0A384AVI1 A0A2F0BBG3 I3MH81 A0A2Y9ETK1 A0A1S3I4B5 K9ISV1 E2RFE5 Q5EA77 A0A1S3AN44 G1NVG7 A0A337SPJ6 A0A2Y9KQN9 A0A3P4S0S0 M3YED2 A0A2K5YKU2 A0A2Y9HMR7 A0A2U3Z2H9 G1L3Y3 H0VAS9 A0A2R5LC33 A0A2K5D0S8 A0A2K6ANR0 A0A2K5X0R5 H2P6N6 Q15036 F6XSD1 A0A182KFR3 F7IBA9
PDB
4TKN
E-value=1.8068e-45,
Score=461
Ontologies
GO
GO:0005623
GO:0035091
GO:0006886
GO:0007165
GO:0005769
GO:1990126
GO:0005856
GO:0008643
GO:0015293
GO:0016021
GO:0030659
GO:0005829
GO:0031410
GO:0006898
GO:0032456
GO:0032991
GO:0010008
GO:0003279
GO:0060976
GO:0005794
GO:0050750
GO:0035904
GO:0008022
GO:0005768
GO:0005737
GO:0030100
GO:0016197
GO:0005102
GO:0016020
GO:0006707
GO:0043231
GO:0006122
GO:0006468
GO:0005524
GO:0004672
GO:0007169
GO:0004713
GO:0006281
GO:0048384
PANTHER
Topology
Subcellular location
Cytoplasm
Early endosome
Cytoplasmic vesicle membrane
Early endosome
Cytoplasmic vesicle membrane
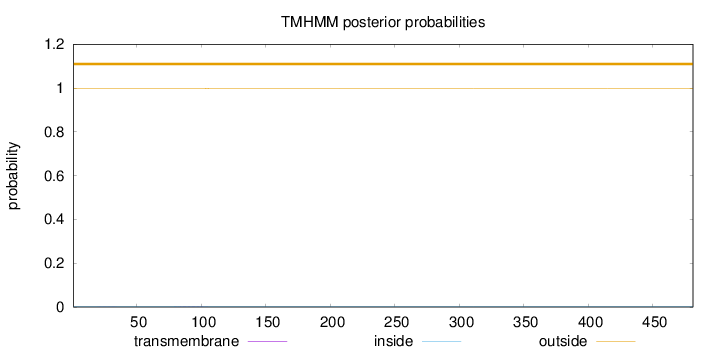
Length:
481
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0342500000000001
Exp number, first 60 AAs:
0.00338
Total prob of N-in:
0.00192
outside
1 - 481
Population Genetic Test Statistics
Pi
266.221327
Theta
194.542443
Tajima's D
1.728163
CLR
0.013505
CSRT
0.836658167091645
Interpretation
Uncertain
