Gene
KWMTBOMO02907 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003544
Annotation
ubiquinol-cytochrome_C_reductase_complex_14kD_subunit_[Bombyx_mori]
Full name
Cytochrome b-c1 complex subunit 7
Location in the cell
Mitochondrial Reliability : 1.773
Sequence
CDS
ATGGCTTTTAGAGCAACTGCCATGATGGCTTTCCGAGCCACGGCTATTAACCGAAGTGACAGCCTCAGCAAATGGGCCTACAATCTTTCGGGATTCAATAAATATGGTTTGTTACGGGATGATTGCTTGCATGAAACTCCTGATGTAACTGAAGCACTCCGCAGACTTCCATCCCATGTTGTTGACGAGAGAAACTTCCGTATTGTACGTGCCATACAGCTCTCCATGCAAAAAACAATCCTACCTAAAGAAGAGTGGACAAAATATGAAGAAGATTCCCTATACTTAACCCCAATTGTTGAGCAAGTTGAGAAAGAGAGGCTGGAGAGAGAGCAGTGGGAGAAGGAATATTAA
Protein
MAFRATAMMAFRATAINRSDSLSKWAYNLSGFNKYGLLRDDCLHETPDVTEALRRLPSHVVDERNFRIVRAIQLSMQKTILPKEEWTKYEEDSLYLTPIVEQVEKERLEREQWEKEY
Summary
Description
Component of the ubiquinol-cytochrome c reductase complex (complex III or cytochrome b-c1 complex), which is part of the mitochondrial respiratory chain.
Similarity
Belongs to the UQCRB/QCR7 family.
Uniprot
Q19P02
S4PGS8
A0A2H1VNP2
A0A1E1WGH8
I4DJH7
A0A0N0PDD4
+ More
A0A0L7LSP0 A0A212F991 H9J207 A0A2W1BRW3 A0A2A4K0G4 A0A0K8SAQ5 A0A1W4XD97 B4MG94 A0A067REU5 R4WPG7 B4PR60 B4QY76 B4IH01 B4L7Q2 B4N931 Q4QPY3 Q9VAW7 A0A034WPK5 A0A0C9RSA0 A0A1L8EHD0 A0A0K8TQ03 A0A1I8Q6Y6 A0A0L0CK99 T1PGX2 A0A182IT82 A0A2S2QQE5 A0A1Q3FNX0 A0A2M3ZFS1 A0A2M4C456 A0A2M4A7W1 A0A182MN12 W5JTE3 T1E8K3 C4WW34 B0WF89 W8C120 D6WQQ1 B3MNX3 A0A0M3QZE8 A0A182U910 Q7Q009 Q5MM88 A0A182W295 A0A182UR69 A0A182JVM6 T1DFM4 C4WRZ3 U5EQ75 A0A023EEP6 A0A182G2V4 A0A1B6CMA0 A0A2H8TJB2 A0A1B6EWG2 A0A1L8E015 A0A0P4ZDH3 A0A0L7R3M4 A0A0M9A6C1 A0A0P5D6Q0 A0A164Z6Q7 A0A0P5ZN65 A0A0P5AER2 A0A0P6E470 A0A0P5QHM3 A0A0P5C852 A0A0P5WVM7 A0A0N8B1I6 A0A0P5LQ03 A0A0N8B498 A0A0P5CPK2 E9GAN1 A0A0P5UR94 A0A0P6G9F6 A0A0P6G4E1 A0A0N7ZMF4 A0A0P5SPY1 A0A182YMJ3 A0A0P5VT97 A0A0P5BJZ3 A0A0P6FP83 A0A0P4YY11 N6STE3 A0A0P5TTD0 A0A0P4ZQJ9 A0A0P6GJK5 A0A0P5BN88 A0A0P4VS46 A0A182PGD9 A0A084WUL9 K7J4C4 A0A171AK51 R4G412 A0A0P5P957 A0A1B0CB78 A0A1B6M2V2
A0A0L7LSP0 A0A212F991 H9J207 A0A2W1BRW3 A0A2A4K0G4 A0A0K8SAQ5 A0A1W4XD97 B4MG94 A0A067REU5 R4WPG7 B4PR60 B4QY76 B4IH01 B4L7Q2 B4N931 Q4QPY3 Q9VAW7 A0A034WPK5 A0A0C9RSA0 A0A1L8EHD0 A0A0K8TQ03 A0A1I8Q6Y6 A0A0L0CK99 T1PGX2 A0A182IT82 A0A2S2QQE5 A0A1Q3FNX0 A0A2M3ZFS1 A0A2M4C456 A0A2M4A7W1 A0A182MN12 W5JTE3 T1E8K3 C4WW34 B0WF89 W8C120 D6WQQ1 B3MNX3 A0A0M3QZE8 A0A182U910 Q7Q009 Q5MM88 A0A182W295 A0A182UR69 A0A182JVM6 T1DFM4 C4WRZ3 U5EQ75 A0A023EEP6 A0A182G2V4 A0A1B6CMA0 A0A2H8TJB2 A0A1B6EWG2 A0A1L8E015 A0A0P4ZDH3 A0A0L7R3M4 A0A0M9A6C1 A0A0P5D6Q0 A0A164Z6Q7 A0A0P5ZN65 A0A0P5AER2 A0A0P6E470 A0A0P5QHM3 A0A0P5C852 A0A0P5WVM7 A0A0N8B1I6 A0A0P5LQ03 A0A0N8B498 A0A0P5CPK2 E9GAN1 A0A0P5UR94 A0A0P6G9F6 A0A0P6G4E1 A0A0N7ZMF4 A0A0P5SPY1 A0A182YMJ3 A0A0P5VT97 A0A0P5BJZ3 A0A0P6FP83 A0A0P4YY11 N6STE3 A0A0P5TTD0 A0A0P4ZQJ9 A0A0P6GJK5 A0A0P5BN88 A0A0P4VS46 A0A182PGD9 A0A084WUL9 K7J4C4 A0A171AK51 R4G412 A0A0P5P957 A0A1B0CB78 A0A1B6M2V2
Pubmed
23622113
22651552
26354079
26227816
22118469
19121390
+ More
28756777 17994087 24845553 23691247 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 26369729 26108605 25315136 20920257 23761445 24495485 18362917 19820115 12364791 14747013 17210077 17510324 24330624 24945155 26483478 21292972 25244985 23537049 27129103 24438588 20075255
28756777 17994087 24845553 23691247 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 26369729 26108605 25315136 20920257 23761445 24495485 18362917 19820115 12364791 14747013 17210077 17510324 24330624 24945155 26483478 21292972 25244985 23537049 27129103 24438588 20075255
EMBL
DQ515929
ABF71568.1
GAIX01006080
JAA86480.1
ODYU01003384
SOQ42072.1
+ More
GDQN01005037 JAT86017.1 AK401445 KQ459597 BAM18067.1 KPI95013.1 KQ460226 KPJ16421.1 JTDY01000168 KOB78472.1 AGBW02009627 OWR50290.1 BABH01007583 KZ149960 PZC76355.1 NWSH01000285 PCG77765.1 GBRD01015459 JAG50367.1 CH940672 EDW57417.1 KK852549 KDR21558.1 AK417546 BAN20761.1 CM000160 EDW98424.1 CM000364 EDX14724.1 CH480837 EDW49119.1 CH933814 EDW05765.1 CH964232 EDW81578.1 BT023633 AAY85033.1 AE014297 KX531464 AAF56781.1 ANY27274.1 GAKP01002827 JAC56125.1 GBYB01010261 JAG80028.1 GFDG01000721 JAV18078.1 GDAI01001382 JAI16221.1 JRES01000292 KNC32685.1 KA647390 AFP62019.1 GGMS01010731 MBY79934.1 GFDL01005821 JAV29224.1 GGFM01006580 MBW27331.1 GGFJ01010933 MBW60074.1 GGFK01003562 MBW36883.1 AXCM01005231 ADMH02000489 ETN66225.1 GAMD01002693 JAA98897.1 ABLF02035906 AK341823 BAH72104.1 DS231915 DS233922 EDS26065.1 EDS32352.1 GAMC01003767 JAC02789.1 KQ971354 EFA07535.1 CH902620 EDV32160.2 CP012528 ALC49249.1 AAAB01008986 EAA00151.4 AY736000 CH477472 AAW21994.1 EAT40390.1 GALA01000522 JAA94330.1 AK339979 BAH70663.1 GANO01003392 JAB56479.1 GAPW01006282 JAC07316.1 JXUM01040150 JXUM01040151 KQ561234 KXJ79276.1 GEDC01022709 JAS14589.1 GFXV01001967 MBW13772.1 GECZ01027443 JAS42326.1 GFDF01001984 JAV12100.1 GDIP01218454 JAJ04948.1 KQ414663 KOC65490.1 KQ435726 KOX78137.1 GDIP01160108 JAJ63294.1 LRGB01000781 KZS16004.1 GDIP01041419 JAM62296.1 GDIP01200756 JAJ22646.1 GDIQ01068728 JAN26009.1 GDIQ01120606 JAL31120.1 GDIP01178354 JAJ45048.1 GDIP01093019 JAM10696.1 GDIP01156355 GDIQ01221971 JAJ67047.1 JAK29754.1 GDIQ01177133 JAK74592.1 GDIQ01214275 JAK37450.1 GDIP01167578 JAJ55824.1 GL732537 EFX83282.1 GDIP01111312 JAL92402.1 GDIQ01036705 JAN58032.1 GDIQ01049585 JAN45152.1 GDIP01231105 JAI92296.1 GDIP01136963 JAL66751.1 GDIP01095668 JAM08047.1 GDIP01183506 JAJ39896.1 GDIQ01044688 JAN50049.1 GDIP01222055 JAJ01347.1 APGK01057490 KB741280 KB632033 ENN70954.1 ERL88175.1 GDIP01123113 JAL80601.1 GDIP01209306 JAJ14096.1 GDIQ01044689 JAN50048.1 GDIP01182307 JAJ41095.1 GDKW01001854 JAI54741.1 ATLV01027100 KE525423 KFB53913.1 AAZX01006683 GEMB01000968 JAS02175.1 ACPB03009391 GAHY01001745 JAA75765.1 GDIQ01153342 JAK98383.1 AJWK01004881 GEBQ01031653 GEBQ01009722 JAT08324.1 JAT30255.1
GDQN01005037 JAT86017.1 AK401445 KQ459597 BAM18067.1 KPI95013.1 KQ460226 KPJ16421.1 JTDY01000168 KOB78472.1 AGBW02009627 OWR50290.1 BABH01007583 KZ149960 PZC76355.1 NWSH01000285 PCG77765.1 GBRD01015459 JAG50367.1 CH940672 EDW57417.1 KK852549 KDR21558.1 AK417546 BAN20761.1 CM000160 EDW98424.1 CM000364 EDX14724.1 CH480837 EDW49119.1 CH933814 EDW05765.1 CH964232 EDW81578.1 BT023633 AAY85033.1 AE014297 KX531464 AAF56781.1 ANY27274.1 GAKP01002827 JAC56125.1 GBYB01010261 JAG80028.1 GFDG01000721 JAV18078.1 GDAI01001382 JAI16221.1 JRES01000292 KNC32685.1 KA647390 AFP62019.1 GGMS01010731 MBY79934.1 GFDL01005821 JAV29224.1 GGFM01006580 MBW27331.1 GGFJ01010933 MBW60074.1 GGFK01003562 MBW36883.1 AXCM01005231 ADMH02000489 ETN66225.1 GAMD01002693 JAA98897.1 ABLF02035906 AK341823 BAH72104.1 DS231915 DS233922 EDS26065.1 EDS32352.1 GAMC01003767 JAC02789.1 KQ971354 EFA07535.1 CH902620 EDV32160.2 CP012528 ALC49249.1 AAAB01008986 EAA00151.4 AY736000 CH477472 AAW21994.1 EAT40390.1 GALA01000522 JAA94330.1 AK339979 BAH70663.1 GANO01003392 JAB56479.1 GAPW01006282 JAC07316.1 JXUM01040150 JXUM01040151 KQ561234 KXJ79276.1 GEDC01022709 JAS14589.1 GFXV01001967 MBW13772.1 GECZ01027443 JAS42326.1 GFDF01001984 JAV12100.1 GDIP01218454 JAJ04948.1 KQ414663 KOC65490.1 KQ435726 KOX78137.1 GDIP01160108 JAJ63294.1 LRGB01000781 KZS16004.1 GDIP01041419 JAM62296.1 GDIP01200756 JAJ22646.1 GDIQ01068728 JAN26009.1 GDIQ01120606 JAL31120.1 GDIP01178354 JAJ45048.1 GDIP01093019 JAM10696.1 GDIP01156355 GDIQ01221971 JAJ67047.1 JAK29754.1 GDIQ01177133 JAK74592.1 GDIQ01214275 JAK37450.1 GDIP01167578 JAJ55824.1 GL732537 EFX83282.1 GDIP01111312 JAL92402.1 GDIQ01036705 JAN58032.1 GDIQ01049585 JAN45152.1 GDIP01231105 JAI92296.1 GDIP01136963 JAL66751.1 GDIP01095668 JAM08047.1 GDIP01183506 JAJ39896.1 GDIQ01044688 JAN50049.1 GDIP01222055 JAJ01347.1 APGK01057490 KB741280 KB632033 ENN70954.1 ERL88175.1 GDIP01123113 JAL80601.1 GDIP01209306 JAJ14096.1 GDIQ01044689 JAN50048.1 GDIP01182307 JAJ41095.1 GDKW01001854 JAI54741.1 ATLV01027100 KE525423 KFB53913.1 AAZX01006683 GEMB01000968 JAS02175.1 ACPB03009391 GAHY01001745 JAA75765.1 GDIQ01153342 JAK98383.1 AJWK01004881 GEBQ01031653 GEBQ01009722 JAT08324.1 JAT30255.1
Proteomes
UP000053268
UP000053240
UP000037510
UP000007151
UP000005204
UP000218220
+ More
UP000192223 UP000008792 UP000027135 UP000002282 UP000000304 UP000001292 UP000009192 UP000007798 UP000000803 UP000095300 UP000037069 UP000095301 UP000075880 UP000075883 UP000000673 UP000007819 UP000002320 UP000007266 UP000007801 UP000092553 UP000075902 UP000007062 UP000008820 UP000075920 UP000075903 UP000075881 UP000069940 UP000249989 UP000053825 UP000053105 UP000076858 UP000000305 UP000076408 UP000019118 UP000030742 UP000075885 UP000030765 UP000002358 UP000015103 UP000092461
UP000192223 UP000008792 UP000027135 UP000002282 UP000000304 UP000001292 UP000009192 UP000007798 UP000000803 UP000095300 UP000037069 UP000095301 UP000075880 UP000075883 UP000000673 UP000007819 UP000002320 UP000007266 UP000007801 UP000092553 UP000075902 UP000007062 UP000008820 UP000075920 UP000075903 UP000075881 UP000069940 UP000249989 UP000053825 UP000053105 UP000076858 UP000000305 UP000076408 UP000019118 UP000030742 UP000075885 UP000030765 UP000002358 UP000015103 UP000092461
Interpro
SUPFAM
SSF81524
SSF81524
Gene 3D
ProteinModelPortal
Q19P02
S4PGS8
A0A2H1VNP2
A0A1E1WGH8
I4DJH7
A0A0N0PDD4
+ More
A0A0L7LSP0 A0A212F991 H9J207 A0A2W1BRW3 A0A2A4K0G4 A0A0K8SAQ5 A0A1W4XD97 B4MG94 A0A067REU5 R4WPG7 B4PR60 B4QY76 B4IH01 B4L7Q2 B4N931 Q4QPY3 Q9VAW7 A0A034WPK5 A0A0C9RSA0 A0A1L8EHD0 A0A0K8TQ03 A0A1I8Q6Y6 A0A0L0CK99 T1PGX2 A0A182IT82 A0A2S2QQE5 A0A1Q3FNX0 A0A2M3ZFS1 A0A2M4C456 A0A2M4A7W1 A0A182MN12 W5JTE3 T1E8K3 C4WW34 B0WF89 W8C120 D6WQQ1 B3MNX3 A0A0M3QZE8 A0A182U910 Q7Q009 Q5MM88 A0A182W295 A0A182UR69 A0A182JVM6 T1DFM4 C4WRZ3 U5EQ75 A0A023EEP6 A0A182G2V4 A0A1B6CMA0 A0A2H8TJB2 A0A1B6EWG2 A0A1L8E015 A0A0P4ZDH3 A0A0L7R3M4 A0A0M9A6C1 A0A0P5D6Q0 A0A164Z6Q7 A0A0P5ZN65 A0A0P5AER2 A0A0P6E470 A0A0P5QHM3 A0A0P5C852 A0A0P5WVM7 A0A0N8B1I6 A0A0P5LQ03 A0A0N8B498 A0A0P5CPK2 E9GAN1 A0A0P5UR94 A0A0P6G9F6 A0A0P6G4E1 A0A0N7ZMF4 A0A0P5SPY1 A0A182YMJ3 A0A0P5VT97 A0A0P5BJZ3 A0A0P6FP83 A0A0P4YY11 N6STE3 A0A0P5TTD0 A0A0P4ZQJ9 A0A0P6GJK5 A0A0P5BN88 A0A0P4VS46 A0A182PGD9 A0A084WUL9 K7J4C4 A0A171AK51 R4G412 A0A0P5P957 A0A1B0CB78 A0A1B6M2V2
A0A0L7LSP0 A0A212F991 H9J207 A0A2W1BRW3 A0A2A4K0G4 A0A0K8SAQ5 A0A1W4XD97 B4MG94 A0A067REU5 R4WPG7 B4PR60 B4QY76 B4IH01 B4L7Q2 B4N931 Q4QPY3 Q9VAW7 A0A034WPK5 A0A0C9RSA0 A0A1L8EHD0 A0A0K8TQ03 A0A1I8Q6Y6 A0A0L0CK99 T1PGX2 A0A182IT82 A0A2S2QQE5 A0A1Q3FNX0 A0A2M3ZFS1 A0A2M4C456 A0A2M4A7W1 A0A182MN12 W5JTE3 T1E8K3 C4WW34 B0WF89 W8C120 D6WQQ1 B3MNX3 A0A0M3QZE8 A0A182U910 Q7Q009 Q5MM88 A0A182W295 A0A182UR69 A0A182JVM6 T1DFM4 C4WRZ3 U5EQ75 A0A023EEP6 A0A182G2V4 A0A1B6CMA0 A0A2H8TJB2 A0A1B6EWG2 A0A1L8E015 A0A0P4ZDH3 A0A0L7R3M4 A0A0M9A6C1 A0A0P5D6Q0 A0A164Z6Q7 A0A0P5ZN65 A0A0P5AER2 A0A0P6E470 A0A0P5QHM3 A0A0P5C852 A0A0P5WVM7 A0A0N8B1I6 A0A0P5LQ03 A0A0N8B498 A0A0P5CPK2 E9GAN1 A0A0P5UR94 A0A0P6G9F6 A0A0P6G4E1 A0A0N7ZMF4 A0A0P5SPY1 A0A182YMJ3 A0A0P5VT97 A0A0P5BJZ3 A0A0P6FP83 A0A0P4YY11 N6STE3 A0A0P5TTD0 A0A0P4ZQJ9 A0A0P6GJK5 A0A0P5BN88 A0A0P4VS46 A0A182PGD9 A0A084WUL9 K7J4C4 A0A171AK51 R4G412 A0A0P5P957 A0A1B0CB78 A0A1B6M2V2
PDB
3H1L
E-value=8.10371e-25,
Score=275
Ontologies
PATHWAY
GO
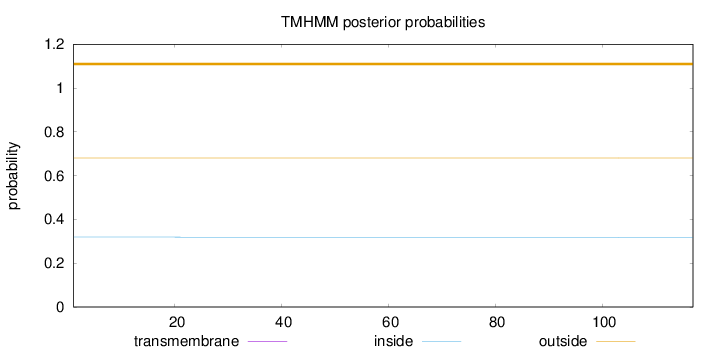
Topology
Subcellular location
Mitochondrion inner membrane
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00073
Exp number, first 60 AAs:
0.00073
Total prob of N-in:
0.31915
outside
1 - 117
Population Genetic Test Statistics
Pi
195.019069
Theta
149.451674
Tajima's D
0.958439
CLR
0
CSRT
0.649167541622919
Interpretation
Uncertain
