Gene
KWMTBOMO02905
Pre Gene Modal
BGIBMGA003646
Annotation
PREDICTED:_uncharacterized_protein_LOC101747221_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.596
Sequence
CDS
ATGGTGAATGGCAACTACGTATTGTATGGGGATAAGCCGTCCTCGGAGTCGGAAATCGTGGATGACCACGTGCATAAACGCACCGTCGACAGAAAAACTAAGCGAAGATCGAGAACAGCTAATGAGCTCCAACGCTCCCTTAATGTGAGGCAACTAGACAATGAAACGGATTCCGTCAAATACGAAAAAAAGATAAACGAACCAAGACTTTTACGGTACGTTCCGACAACTGATGTAGAAGAAAATTATGTGACAGAAAAATTCGAAACCTATGATTTGAAGCCGACCGAAGAACGACCTAGAAAATACGCATTTACTAAGAAACCTAAGAACCCAACGGATGGCTCTATTAATTACGCGTATTCGAGGTCAAGCAGTAGTTGTAGTTCTCCAAACGGAAATTTACCGACCATATCGGAGCACGGAGAACAGTACGGGAATTATCGAGTCCATTCTTTTTCGGGTGCCGTGATTGATGATGTCGTCACCGGCTACGAGACGAAACTTGATAAATACTTCTCCAAGCCGCGGGAAGTCAAGACATTTGCTGCTAAGGGATCTCACCCAGACAGCCTGTACAGAGTTCCAGTTGACAGTGGAGTCGCACGTACTAAGATGACAAGAGTGGTACGAGTTATCCGATGGCCCGCTGCCCTTGTAGCCGTATGCGTCGCCTTAGCAGTCTTCGTGTACTTCCTTATGCCTGATACGATCGACATTGATGTAGACTCTGTTAATGGAACTACTTGGGAAGCTGCGAGAGCACACAATAATGTATTACCTCACAAATTTGAAGAGAGTAAAGTCAAGAAGAATGACAATGATCGTGCTCAAACACAGGACTTTTACGATGCTGATAATGACAAGAGTTACGATATTACCGCTATAATTGATACAATTACGGAAACGGATCCTAAAAGGGTGCTGCCGGTTGCACCAATATTCCCGAGTCATATCACTCCAGAGGTGCAATACGGGAATGAAAGGGTTGATAAAGATAAATTGAGAACGCCGAAAGTAATTAACGAAACGAACACATTTCACGAAGACTCAACACTCAAACCACATATCACTCAAAAACCTGAAAAACCCTTAGAAGTATACTTTAGGGACGTCACCGCAATCCCTACAACCACGGAATCTATCTTCAGAGCTGAAACGGTTAACGATTTTACGAATTACGAAGAGAAACAAACCACGAAAAAGATCGAAGAAGCGTCTTCGAAACATCGCCCATCACTTTACAAACAAAACTTCGTTCAACTTCCAAACGGAGTACAAGATGTCGAAGCGTCATCCGAGATACCTCTCGAAGTGCAAGAATACTACGGCGAGCAACCGGTGAAATCTAATGTTAACTTTACGTCTGGTCACTCGAAATTGTTCGGCATTTCTATGGAAGATGCGGAAAAAATTAAATCTACCACGCAAAGTTCAGTGTACAACACTAGGGTCTCACCCACGCTTCCGACTTGGCGGGATGGTGACGACGCTACTACCAAAAAATATCCAACTAACATTAATTACGAAGTGCCGCAGTGTCATTCGACCCACATACCTCTGTGCCGTGGAGTGCTGCCCTACGACTTAGCAGGAAACGCAGCTTCAGTCGGTGGAATAGAGATCACGGCCTTACTACCGCAGATAGAATACTTGGTGGCCACTAACTGCAGCGACCGTGTGAGGCACTTTGTCTGCGCTCTACTCGAACCTGAATGCAACCCGCCTCCGTATCCACCGAAATTGCCATGCTTTAATCTTTGCAAAGCTATCGTTGACAACTGTGACGGGCTGATCCCCCGAGACCTATCCCCGGCGTTCAATTGTCAGCAATATTTCAAGAACAACTGTATAGCAGCCAAGACGCCGTGCTACCCGAGAGAGATGACCTGCGGCGACGGATCCTGCATCCCGAGGGACTGGATCTGCGACGGGACCCGGGACTGCCTCAGCGGAGAAGACGAGGCGACTTGCGTCCAATGCGACACCCGCGAGTTCCGGTGTCCGTCCGGGGGATGTATACAGAAGCGTTGGCTCTGCGACGGGTACGCGGACTGCTCTAACGGGGAAGACGAGGACGTCGAGGTGTGCGGCAGCCCCGTGGTGGCGGGCGAGGAGCCGGCCGGGTCCGCGCCCGCACCCGCCGTGCGCAGACCGAACAGAATGCAGAATTCACACGGACGACAATACTCACACAATACAGGTGAGAATGAAAGCAGCAAAGAGCTCTTGGTCACAAGCGACAGCAGCAACTCCCTCAAACGGAATTTCACACGAAGACCCTCGCTGTCCCGACTGACGCCCTACAACCGAAATATGCCGCATACAATCCAAACCAGACATAACAATACACAAACAACCACAGCTCCCGTCACCAAACACTCCTCGCAAGACAACGAGAGAACAACACAGAACGCTGACGATGGAGATTCCGAGGAAGACGTGAATGTCGGAGACCTCGGATTCTTCGAAGACCTGAAGAAAGAACCTAAAGAACAGCCCGCTAAGCGGGTGATTGCGAAATCGCAGATGACCAAACCGAAGTCGTCTATAGAAAAAGACGCGGGAGGCGGCGCCCCGGCCCCCACGCGACTCGATCAGTCCATCAGCAAACTGGAGAGGGTTATCAATGGAGCGGCTCTACTAAAGAAAGCGGAAGCCGAATCGAAATCGGAAAAGGCGGAAGAGTTAGAAGAATCGCCTGAAATGAATAAGGGCGGGACGAGGAGTGACGATCTGCCGACCGGACCGCGGGGAGGGTCGACCAGCGCGCATGTGTCGCCCTGTCCCACCGGGGAGTTACGCTGCGTGGACGGGCGCTGCATCACGCTGGCCCAACTCTGCGACGGCACCATCGACTGCTCCGACCACGCAGACGAAGACAACTGCTTCACGTGA
Protein
MVNGNYVLYGDKPSSESEIVDDHVHKRTVDRKTKRRSRTANELQRSLNVRQLDNETDSVKYEKKINEPRLLRYVPTTDVEENYVTEKFETYDLKPTEERPRKYAFTKKPKNPTDGSINYAYSRSSSSCSSPNGNLPTISEHGEQYGNYRVHSFSGAVIDDVVTGYETKLDKYFSKPREVKTFAAKGSHPDSLYRVPVDSGVARTKMTRVVRVIRWPAALVAVCVALAVFVYFLMPDTIDIDVDSVNGTTWEAARAHNNVLPHKFEESKVKKNDNDRAQTQDFYDADNDKSYDITAIIDTITETDPKRVLPVAPIFPSHITPEVQYGNERVDKDKLRTPKVINETNTFHEDSTLKPHITQKPEKPLEVYFRDVTAIPTTTESIFRAETVNDFTNYEEKQTTKKIEEASSKHRPSLYKQNFVQLPNGVQDVEASSEIPLEVQEYYGEQPVKSNVNFTSGHSKLFGISMEDAEKIKSTTQSSVYNTRVSPTLPTWRDGDDATTKKYPTNINYEVPQCHSTHIPLCRGVLPYDLAGNAASVGGIEITALLPQIEYLVATNCSDRVRHFVCALLEPECNPPPYPPKLPCFNLCKAIVDNCDGLIPRDLSPAFNCQQYFKNNCIAAKTPCYPREMTCGDGSCIPRDWICDGTRDCLSGEDEATCVQCDTREFRCPSGGCIQKRWLCDGYADCSNGEDEDVEVCGSPVVAGEEPAGSAPAPAVRRPNRMQNSHGRQYSHNTGENESSKELLVTSDSSNSLKRNFTRRPSLSRLTPYNRNMPHTIQTRHNNTQTTTAPVTKHSSQDNERTTQNADDGDSEEDVNVGDLGFFEDLKKEPKEQPAKRVIAKSQMTKPKSSIEKDAGGGAPAPTRLDQSISKLERVINGAALLKKAEAESKSEKAEELEESPEMNKGGTRSDDLPTGPRGGSTSAHVSPCPTGELRCVDGRCITLAQLCDGTIDCSDHADEDNCFT
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
PDB
1N7D
E-value=1.17486e-13,
Score=190
Ontologies
GO
PANTHER
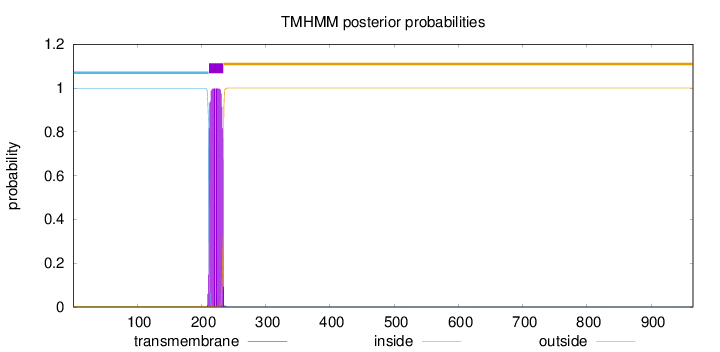
Topology
Length:
965
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.32216
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99675
inside
1 - 211
TMhelix
212 - 234
outside
235 - 965
Population Genetic Test Statistics
Pi
223.705733
Theta
171.137039
Tajima's D
0.955039
CLR
0.339843
CSRT
0.651617419129044
Interpretation
Uncertain
