Pre Gene Modal
BGIBMGA003648
Annotation
PREDICTED:_ubiquitin-protein_ligase_E3C_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.975
Sequence
CDS
ATGGCGTACGTGTGCACGCACCTGCTGCGTGCGCTGTACGTGCGCGACTCGCGGCTGCAGTTCTGCGGCAGCGCGGCGTGGCCGGTGGAGGGCGCCATCCCCGAGGGGGGCGGGGCCGCGCTCGTCACGCACGCTGTGCACTCGCGACCCTTCCCCACGCATCTGCGGCAGCTGAACCTCTCGCAAGAAGAGGGCCCGCCGCTGACAATCAAGGAGCTGAGAACAGTCACGATACTCAAAGAAATACCATTCGTGGTGCCGTTCTCAACGCGAGTCCTGATATTCCAAGCGCTACTCGCAAAAGAAAAACACGACAATTGGTATGAGATGACTAATTTCAGTGAAGGTCCGTTTATAAACATTAGCGTCCGACGCACGCATTTGTATGAAGACGCTTTCGACAAACTCAGTCCCGATAATGAACCAAATATGAAATTAAAGTTACGAGTACAACTGATCAATCAAGCCGGCGCCGAAGAGGCTGGCGTCGACGGTGGAGGCCTATTCCGAGAATTCATATCCGAATTACTTAAATCAGCGTTTGATCCAAACAGAGGGTTGTTTCGTTTAACGAAAGATAATATGCTGTACCCGAATCCTGGCGTTAACTTGTTGTACGATGATTTTCCCATGCACTATTACTTCGTCGGCAGGATGCTTGGAAAGGCACTGTACGAAAATTTACTGGTAGAACTTCCACTAGCGGAGTTTTTCCTCGCCAAGCTGTGCTCGTCTCGGGAGCCGGACGTTCACGCCTTGGCCTCTTTGGATCCAGGCTTGTACCGGGGGCTATTGATGCTGAAGTCTCATCGAAGGCAAGACGTTCCGGAGCTGGGACTCGACTTCACGATACTCAGCGATGAACTGGGCGAGCAAAGGATAGATGAACTGAAACCCGGCGGTGCCAACATTCCAGTGACCGCTGAAAATAGGATAGAATATATTCATCTAGTCGCCGATTATAAATTGAACAGACAAATAAGGTCTCAGTGCATCGCGTTCAAGCGCGGGCTGGCGTCGGTCGTGAACGCGGACTGGCTGCGGATGTTCTCGTGCCGGGAGCTGCAGCTGCTGGTGTCGGGCGCCGAGGTGCCCATCGACGTGCGCGACCTCCGACAACACACGCTCTACTCCGGAGGCTTTTCGGATTCGCATCCCACCGTGCAGTGCTTCTGGAAGGTGGTCGAGAACCTCACGGATGACCAGAGACGACAGCTGCTGAAATTCGTCACGAGCTGTTCGCGGCCTCCCTTGTTGGGTTTTAAGGACCTGCAGCCCCCGTTCTGCATACAGTCTGCGGGTGCGTCGGATCGACTCCCGTCGTCGTCGACCTGCATGAACCTGCTGAAGCTGCCGGAGTTCCACAGCGAGGAGCAGCTCGCGGAGAAGTTACTGTACGCGATACAGGCCGGGGCCGGATTCGAGCTCAGCTAA
Protein
MAYVCTHLLRALYVRDSRLQFCGSAAWPVEGAIPEGGGAALVTHAVHSRPFPTHLRQLNLSQEEGPPLTIKELRTVTILKEIPFVVPFSTRVLIFQALLAKEKHDNWYEMTNFSEGPFINISVRRTHLYEDAFDKLSPDNEPNMKLKLRVQLINQAGAEEAGVDGGGLFREFISELLKSAFDPNRGLFRLTKDNMLYPNPGVNLLYDDFPMHYYFVGRMLGKALYENLLVELPLAEFFLAKLCSSREPDVHALASLDPGLYRGLLMLKSHRRQDVPELGLDFTILSDELGEQRIDELKPGGANIPVTAENRIEYIHLVADYKLNRQIRSQCIAFKRGLASVVNADWLRMFSCRELQLLVSGAEVPIDVRDLRQHTLYSGGFSDSHPTVQCFWKVVENLTDDQRRQLLKFVTSCSRPPLLGFKDLQPPFCIQSAGASDRLPSSSTCMNLLKLPEFHSEEQLAEKLLYAIQAGAGFELS
Summary
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Uniprot
H9J2B1
A0A1E1WBV5
A0A1E1WF09
A0A2A4K259
A0A2H1WDG2
A0A2W1BQ10
+ More
A0A0N1IFM7 A0A194PV63 A0A212ELL8 S4PIZ6 K7IPJ0 A0A087ZWH0 A0A232F3V6 A0A2A3EMP7 A0A0M9A9L3 A0A151JTB7 A0A026WIC3 A0A0L7QV74 F4WYP1 E9J4K9 A0A158NM58 A0A195BXQ3 A0A151J589 A0A067QLC7 A0A154PTM6 A0A0C9RNM0 E2A6Z2 A0A151IE76 A0A0C9RXK1 E2C2T7 A0A2J7PQJ9 A0A2J7PQJ8 A0A2J7PQJ1 A0A151XJL4 A0A1B6EYS1 E0VR04 A0A1B6CRY1 A0A1Q3F600 B0W765 A0A1Q3F789 A0A182GYL8 A0A1S4FTM8 A0A0P6IYF2 Q16PR2 Q16J44 A0A182ILF2 A0A182MXZ7 A0A182QT97 A0A084WBD0 A0A182R8P1 A0A182WF68 A0A182YPE4 A0A182FPN8 A0A336MWB4 A0A2M4AIX8 W5JV43 A0A182P9B5 A0A336MW41 A0A182XC90 Q7Q342 A0A0P5DWD5 A0A0P5CVL5 A0A0P4YCM7 A0A0P5DE48 A0A0P5UBL6 A0A0P4WL61 A0A0P4WPH6 A0A0P5D6X4 A0A0P5ALZ8 A0A0P6C6A5 A0A0N8AC83 A0A0P5WEN9 A0A0P5EA89 A0A0P4YUI5 E9GES0 A0A182JPA3 A0A0P6IG15 T1J5A0 A0A0N8BTT7 A0A1J1I3Y3 A0A2S2PNG9 A0A182MEI8 A0A2S2QMC3 A0A0N8B6H1 A0A2H8TTS2 A0A293L7M4 A0A2S2PWD8 J9JJG6 A0A2T7NQH5 A0A0N8AFX1 T1K5B5 A0A0P5SUG1 K1QNF6 R7TFZ0 A0A1S3JYX9 V4B5L0 Q9W196 B3NQG8 B4I2F2 A0A0A1XNT7
A0A0N1IFM7 A0A194PV63 A0A212ELL8 S4PIZ6 K7IPJ0 A0A087ZWH0 A0A232F3V6 A0A2A3EMP7 A0A0M9A9L3 A0A151JTB7 A0A026WIC3 A0A0L7QV74 F4WYP1 E9J4K9 A0A158NM58 A0A195BXQ3 A0A151J589 A0A067QLC7 A0A154PTM6 A0A0C9RNM0 E2A6Z2 A0A151IE76 A0A0C9RXK1 E2C2T7 A0A2J7PQJ9 A0A2J7PQJ8 A0A2J7PQJ1 A0A151XJL4 A0A1B6EYS1 E0VR04 A0A1B6CRY1 A0A1Q3F600 B0W765 A0A1Q3F789 A0A182GYL8 A0A1S4FTM8 A0A0P6IYF2 Q16PR2 Q16J44 A0A182ILF2 A0A182MXZ7 A0A182QT97 A0A084WBD0 A0A182R8P1 A0A182WF68 A0A182YPE4 A0A182FPN8 A0A336MWB4 A0A2M4AIX8 W5JV43 A0A182P9B5 A0A336MW41 A0A182XC90 Q7Q342 A0A0P5DWD5 A0A0P5CVL5 A0A0P4YCM7 A0A0P5DE48 A0A0P5UBL6 A0A0P4WL61 A0A0P4WPH6 A0A0P5D6X4 A0A0P5ALZ8 A0A0P6C6A5 A0A0N8AC83 A0A0P5WEN9 A0A0P5EA89 A0A0P4YUI5 E9GES0 A0A182JPA3 A0A0P6IG15 T1J5A0 A0A0N8BTT7 A0A1J1I3Y3 A0A2S2PNG9 A0A182MEI8 A0A2S2QMC3 A0A0N8B6H1 A0A2H8TTS2 A0A293L7M4 A0A2S2PWD8 J9JJG6 A0A2T7NQH5 A0A0N8AFX1 T1K5B5 A0A0P5SUG1 K1QNF6 R7TFZ0 A0A1S3JYX9 V4B5L0 Q9W196 B3NQG8 B4I2F2 A0A0A1XNT7
Pubmed
19121390
28756777
26354079
22118469
23622113
20075255
+ More
28648823 24508170 30249741 21719571 21282665 21347285 24845553 20798317 20566863 26483478 26999592 17510324 24438588 25244985 20920257 23761445 12364791 21292972 22992520 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 25830018
28648823 24508170 30249741 21719571 21282665 21347285 24845553 20798317 20566863 26483478 26999592 17510324 24438588 25244985 20920257 23761445 12364791 21292972 22992520 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 25830018
EMBL
BABH01007596
BABH01007597
BABH01007598
BABH01007599
BABH01007600
GDQN01006625
+ More
JAT84429.1 GDQN01005476 JAT85578.1 NWSH01000285 PCG77760.1 ODYU01007914 SOQ51109.1 KZ149960 PZC76361.1 KQ460226 KPJ16427.1 KQ459597 KPI95020.1 AGBW02014037 OWR42368.1 GAIX01005225 JAA87335.1 NNAY01001008 OXU25516.1 KZ288215 PBC32476.1 KQ435713 KOX79296.1 KQ981960 KYN31906.1 KK107238 QOIP01000007 EZA54854.1 RLU20360.1 KQ414727 KOC62552.1 GL888455 EGI60676.1 GL768105 EFZ12282.1 ADTU01020292 KQ976395 KYM93075.1 KQ980039 KYN18133.1 KK853201 KDR09906.1 KQ435127 KZC14784.1 GBYB01010005 JAG79772.1 GL437208 EFN70795.1 KQ977890 KYM98999.1 GBYB01013895 JAG83662.1 GL452203 EFN77772.1 NEVH01022635 PNF18607.1 PNF18609.1 PNF18610.1 KQ982060 KYQ60579.1 GECZ01026610 JAS43159.1 DS235442 EEB15810.1 GEDC01021120 JAS16178.1 GFDL01012050 JAV22995.1 DS231852 EDS37670.1 GFDL01011697 JAV23348.1 JXUM01021198 JXUM01021199 KQ560594 KXJ81712.1 GDUN01001031 JAN94888.1 CH477774 EAT36361.1 CH478032 EAT34283.1 AXCN02001648 ATLV01022343 KE525331 KFB47524.1 UFQT01002994 SSX34410.1 GGFK01007428 MBW40749.1 ADMH02000409 ETN66654.1 UFQS01003124 UFQT01003124 SSX15237.1 SSX34612.1 AAAB01008966 EAA13012.4 GDIP01156480 JAJ66922.1 GDIP01181116 JAJ42286.1 GDIP01239072 JAI84329.1 GDIP01158872 JAJ64530.1 GDIP01115370 JAL88344.1 GDIP01254756 JAI68645.1 GDIP01254757 JAI68644.1 GDIP01160035 JAJ63367.1 GDIP01196920 JAJ26482.1 GDIP01020361 JAM83354.1 GDIP01161673 JAJ61729.1 GDIP01087181 JAM16534.1 GDIP01146027 JAJ77375.1 GDIP01222518 JAJ00884.1 GL732541 EFX82031.1 GDIQ01006569 JAN88168.1 JH431855 GDIQ01145563 JAL06163.1 CVRI01000040 CRK94907.1 GGMR01018318 MBY30937.1 AXCM01000254 GGMS01009099 MBY78302.1 GDIQ01208091 JAK43634.1 GFXV01005821 MBW17626.1 GFWV01003967 MAA28697.1 GGMS01000568 MBY69771.1 ABLF02033481 ABLF02033487 PZQS01000010 PVD23402.1 GDIP01151369 JAJ72033.1 CAEY01001585 GDIP01135125 JAL68589.1 JH815790 EKC38417.1 AMQN01013239 KB310056 ELT92644.1 KB200521 ESP01317.1 AE013599 AY069726 AAF47181.1 AAL39871.1 CH954179 EDV56971.1 CH480820 EDW53947.1 GBXI01001742 JAD12550.1
JAT84429.1 GDQN01005476 JAT85578.1 NWSH01000285 PCG77760.1 ODYU01007914 SOQ51109.1 KZ149960 PZC76361.1 KQ460226 KPJ16427.1 KQ459597 KPI95020.1 AGBW02014037 OWR42368.1 GAIX01005225 JAA87335.1 NNAY01001008 OXU25516.1 KZ288215 PBC32476.1 KQ435713 KOX79296.1 KQ981960 KYN31906.1 KK107238 QOIP01000007 EZA54854.1 RLU20360.1 KQ414727 KOC62552.1 GL888455 EGI60676.1 GL768105 EFZ12282.1 ADTU01020292 KQ976395 KYM93075.1 KQ980039 KYN18133.1 KK853201 KDR09906.1 KQ435127 KZC14784.1 GBYB01010005 JAG79772.1 GL437208 EFN70795.1 KQ977890 KYM98999.1 GBYB01013895 JAG83662.1 GL452203 EFN77772.1 NEVH01022635 PNF18607.1 PNF18609.1 PNF18610.1 KQ982060 KYQ60579.1 GECZ01026610 JAS43159.1 DS235442 EEB15810.1 GEDC01021120 JAS16178.1 GFDL01012050 JAV22995.1 DS231852 EDS37670.1 GFDL01011697 JAV23348.1 JXUM01021198 JXUM01021199 KQ560594 KXJ81712.1 GDUN01001031 JAN94888.1 CH477774 EAT36361.1 CH478032 EAT34283.1 AXCN02001648 ATLV01022343 KE525331 KFB47524.1 UFQT01002994 SSX34410.1 GGFK01007428 MBW40749.1 ADMH02000409 ETN66654.1 UFQS01003124 UFQT01003124 SSX15237.1 SSX34612.1 AAAB01008966 EAA13012.4 GDIP01156480 JAJ66922.1 GDIP01181116 JAJ42286.1 GDIP01239072 JAI84329.1 GDIP01158872 JAJ64530.1 GDIP01115370 JAL88344.1 GDIP01254756 JAI68645.1 GDIP01254757 JAI68644.1 GDIP01160035 JAJ63367.1 GDIP01196920 JAJ26482.1 GDIP01020361 JAM83354.1 GDIP01161673 JAJ61729.1 GDIP01087181 JAM16534.1 GDIP01146027 JAJ77375.1 GDIP01222518 JAJ00884.1 GL732541 EFX82031.1 GDIQ01006569 JAN88168.1 JH431855 GDIQ01145563 JAL06163.1 CVRI01000040 CRK94907.1 GGMR01018318 MBY30937.1 AXCM01000254 GGMS01009099 MBY78302.1 GDIQ01208091 JAK43634.1 GFXV01005821 MBW17626.1 GFWV01003967 MAA28697.1 GGMS01000568 MBY69771.1 ABLF02033481 ABLF02033487 PZQS01000010 PVD23402.1 GDIP01151369 JAJ72033.1 CAEY01001585 GDIP01135125 JAL68589.1 JH815790 EKC38417.1 AMQN01013239 KB310056 ELT92644.1 KB200521 ESP01317.1 AE013599 AY069726 AAF47181.1 AAL39871.1 CH954179 EDV56971.1 CH480820 EDW53947.1 GBXI01001742 JAD12550.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000002358
+ More
UP000005203 UP000215335 UP000242457 UP000053105 UP000078541 UP000053097 UP000279307 UP000053825 UP000007755 UP000005205 UP000078540 UP000078492 UP000027135 UP000076502 UP000000311 UP000078542 UP000008237 UP000235965 UP000075809 UP000009046 UP000002320 UP000069940 UP000249989 UP000008820 UP000075880 UP000075884 UP000075886 UP000030765 UP000075900 UP000075920 UP000076408 UP000069272 UP000000673 UP000075885 UP000076407 UP000007062 UP000000305 UP000075881 UP000183832 UP000075883 UP000007819 UP000245119 UP000015104 UP000005408 UP000014760 UP000085678 UP000030746 UP000000803 UP000008711 UP000001292
UP000005203 UP000215335 UP000242457 UP000053105 UP000078541 UP000053097 UP000279307 UP000053825 UP000007755 UP000005205 UP000078540 UP000078492 UP000027135 UP000076502 UP000000311 UP000078542 UP000008237 UP000235965 UP000075809 UP000009046 UP000002320 UP000069940 UP000249989 UP000008820 UP000075880 UP000075884 UP000075886 UP000030765 UP000075900 UP000075920 UP000076408 UP000069272 UP000000673 UP000075885 UP000076407 UP000007062 UP000000305 UP000075881 UP000183832 UP000075883 UP000007819 UP000245119 UP000015104 UP000005408 UP000014760 UP000085678 UP000030746 UP000000803 UP000008711 UP000001292
Interpro
CDD
ProteinModelPortal
H9J2B1
A0A1E1WBV5
A0A1E1WF09
A0A2A4K259
A0A2H1WDG2
A0A2W1BQ10
+ More
A0A0N1IFM7 A0A194PV63 A0A212ELL8 S4PIZ6 K7IPJ0 A0A087ZWH0 A0A232F3V6 A0A2A3EMP7 A0A0M9A9L3 A0A151JTB7 A0A026WIC3 A0A0L7QV74 F4WYP1 E9J4K9 A0A158NM58 A0A195BXQ3 A0A151J589 A0A067QLC7 A0A154PTM6 A0A0C9RNM0 E2A6Z2 A0A151IE76 A0A0C9RXK1 E2C2T7 A0A2J7PQJ9 A0A2J7PQJ8 A0A2J7PQJ1 A0A151XJL4 A0A1B6EYS1 E0VR04 A0A1B6CRY1 A0A1Q3F600 B0W765 A0A1Q3F789 A0A182GYL8 A0A1S4FTM8 A0A0P6IYF2 Q16PR2 Q16J44 A0A182ILF2 A0A182MXZ7 A0A182QT97 A0A084WBD0 A0A182R8P1 A0A182WF68 A0A182YPE4 A0A182FPN8 A0A336MWB4 A0A2M4AIX8 W5JV43 A0A182P9B5 A0A336MW41 A0A182XC90 Q7Q342 A0A0P5DWD5 A0A0P5CVL5 A0A0P4YCM7 A0A0P5DE48 A0A0P5UBL6 A0A0P4WL61 A0A0P4WPH6 A0A0P5D6X4 A0A0P5ALZ8 A0A0P6C6A5 A0A0N8AC83 A0A0P5WEN9 A0A0P5EA89 A0A0P4YUI5 E9GES0 A0A182JPA3 A0A0P6IG15 T1J5A0 A0A0N8BTT7 A0A1J1I3Y3 A0A2S2PNG9 A0A182MEI8 A0A2S2QMC3 A0A0N8B6H1 A0A2H8TTS2 A0A293L7M4 A0A2S2PWD8 J9JJG6 A0A2T7NQH5 A0A0N8AFX1 T1K5B5 A0A0P5SUG1 K1QNF6 R7TFZ0 A0A1S3JYX9 V4B5L0 Q9W196 B3NQG8 B4I2F2 A0A0A1XNT7
A0A0N1IFM7 A0A194PV63 A0A212ELL8 S4PIZ6 K7IPJ0 A0A087ZWH0 A0A232F3V6 A0A2A3EMP7 A0A0M9A9L3 A0A151JTB7 A0A026WIC3 A0A0L7QV74 F4WYP1 E9J4K9 A0A158NM58 A0A195BXQ3 A0A151J589 A0A067QLC7 A0A154PTM6 A0A0C9RNM0 E2A6Z2 A0A151IE76 A0A0C9RXK1 E2C2T7 A0A2J7PQJ9 A0A2J7PQJ8 A0A2J7PQJ1 A0A151XJL4 A0A1B6EYS1 E0VR04 A0A1B6CRY1 A0A1Q3F600 B0W765 A0A1Q3F789 A0A182GYL8 A0A1S4FTM8 A0A0P6IYF2 Q16PR2 Q16J44 A0A182ILF2 A0A182MXZ7 A0A182QT97 A0A084WBD0 A0A182R8P1 A0A182WF68 A0A182YPE4 A0A182FPN8 A0A336MWB4 A0A2M4AIX8 W5JV43 A0A182P9B5 A0A336MW41 A0A182XC90 Q7Q342 A0A0P5DWD5 A0A0P5CVL5 A0A0P4YCM7 A0A0P5DE48 A0A0P5UBL6 A0A0P4WL61 A0A0P4WPH6 A0A0P5D6X4 A0A0P5ALZ8 A0A0P6C6A5 A0A0N8AC83 A0A0P5WEN9 A0A0P5EA89 A0A0P4YUI5 E9GES0 A0A182JPA3 A0A0P6IG15 T1J5A0 A0A0N8BTT7 A0A1J1I3Y3 A0A2S2PNG9 A0A182MEI8 A0A2S2QMC3 A0A0N8B6H1 A0A2H8TTS2 A0A293L7M4 A0A2S2PWD8 J9JJG6 A0A2T7NQH5 A0A0N8AFX1 T1K5B5 A0A0P5SUG1 K1QNF6 R7TFZ0 A0A1S3JYX9 V4B5L0 Q9W196 B3NQG8 B4I2F2 A0A0A1XNT7
PDB
5XMC
E-value=6.30376e-43,
Score=439
Ontologies
KEGG
GO
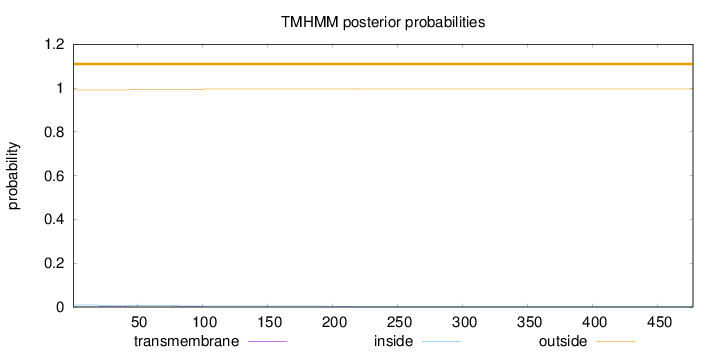
Topology
Length:
477
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11689
Exp number, first 60 AAs:
0.0466
Total prob of N-in:
0.00916
outside
1 - 477
Population Genetic Test Statistics
Pi
230.756703
Theta
163.980688
Tajima's D
1.349722
CLR
0.005204
CSRT
0.752712364381781
Interpretation
Uncertain
