Gene
KWMTBOMO02897
Pre Gene Modal
BGIBMGA003650
Annotation
PREDICTED:_voltage-dependent_calcium_channel_subunit_alpha-2/delta-3_[Bombyx_mori]
Location in the cell
Lysosomal Reliability : 1.701
Sequence
CDS
ATGTATAAGCTTGCGGATAATCAAAGCAGGGTCGACACGCAACCATTGAGAATACCAGCGAACCGAAAGGGTTGCCAAATGTCACCTCACAAGCGGATCATGGACAGCGCCGAAGCCGCCGCGCTATCATCAAGCAGTGGATCTGAAGGCACCCCCGGTACATATTACGACGCGCGATGGTTAAATGTCCACGCTGATGATGGAACTCTGGCGGCGAAGGCACGACGTTTGCTCCTCGCCCCCAGTCGGCACTTTGATCACATCGCTGTCAACACTAGCTATAGTGCTGTTTTGATGCCGCCCTATATCAGCGTTGAAGATCCCGAAGTGCAAAATCAGATCGCGTGGTCAGAGCACTTGGATCCTTTGTTCGTTAACAACTACGAAATAGATCCTACATTGTCGTGGCAATATTATGCTTCTTCCAGTGGATTTATGAGGAGATATCCAGCAATGTCCTGGCCTCCGGAAGACGGCTATTCACACCACGCCCGAGACTTTTACGACTTCAGATCGTCGAACTGGTTCGTCGAGTCGGCGACATCTCCGAAAGACTTGGTCATTCTACTCGATGACTCTGATGACGTCAGCTCATCGTACTGGAGACTGGCGAAGGCTACCGTATCGGCTCTACTCGACACGCTCGGCCCTAACGACTTCGTGAATGTGTACCGTTACAGCGACACGGTAGCGGAGATAAACTCTTGCTACACGAAGGTTTTAGCTCAGGCCCTCCCTGAAACCCTGCGTGAAATGAAAACGTCGTTGTGGAATGCCGAAATAACTGGCGGAATCGCGAACATAACAGGGGCCCTCACCACAGCCTTCGAAATTCTACATCGGTATAATCGTACCGGACAAGGGTGCCAGTGCAACCAGGCTATAGCGGTGGTGGGGGCGGGCGGCGGCGCTGGTGGCGTCAACCGCGTGTTCCGCACTTGGAACTGGCCGCACATGCCCGTCAGAATATTTACGTACCGAACGGGAGGAGACGCGGCTGCCGGGGTATATATGCAGGAAATGGCATGCACAAACAAAGGTTTCTACGTAACTATAAACGACCACTCCGAGATTCGGCCAAAAGTTCTTCACTTCGTCGAAGTACTGGCGCGTCCGCTCGTCATGTACCAAAATGTCCACCCGGTGCACTGGAGTCCGGTTTACGTCGGAGGACGAAGCAGTAGCCTCGCCGATAACGGAATGGGTCAGCTTATGACATCAGTAACAGTTCCCATATTCGACAGGAGAAACCATACGCAAAGGGAAGCGAATTTACTAGGTGCTGTTGGAACAGACGTGCCGGTCGATCAAATAAAGAAGCTCGTACCACCGTACAAGTTGGGCGTTAATGGCTATTCGTTCATGGTGGACAACAACGGGCATGTATTATACCACCCGGACTTAAGACCATTGCATACAAACGAAGAAGTAAGTAGAATGTTAATCTAA
Protein
MYKLADNQSRVDTQPLRIPANRKGCQMSPHKRIMDSAEAAALSSSSGSEGTPGTYYDARWLNVHADDGTLAAKARRLLLAPSRHFDHIAVNTSYSAVLMPPYISVEDPEVQNQIAWSEHLDPLFVNNYEIDPTLSWQYYASSSGFMRRYPAMSWPPEDGYSHHARDFYDFRSSNWFVESATSPKDLVILLDDSDDVSSSYWRLAKATVSALLDTLGPNDFVNVYRYSDTVAEINSCYTKVLAQALPETLREMKTSLWNAEITGGIANITGALTTAFEILHRYNRTGQGCQCNQAIAVVGAGGGAGGVNRVFRTWNWPHMPVRIFTYRTGGDAAAGVYMQEMACTNKGFYVTINDHSEIRPKVLHFVEVLARPLVMYQNVHPVHWSPVYVGGRSSSLADNGMGQLMTSVTVPIFDRRNHTQREANLLGAVGTDVPVDQIKKLVPPYKLGVNGYSFMVDNNGHVLYHPDLRPLHTNEEVSRMLI
Summary
Uniprot
H9J2B3
A0A212ELK1
A0A2W1BUN7
A0A194PNL7
A0A0N0PDD7
A0A2P8XES1
+ More
A0A067RSJ0 A0A1W4WYM4 D7EJQ5 A0A139WP66 A0A1W4WNW6 A0A1Y1NDS7 A0A151IZ18 A0A0N0BJP4 A0A3L8D6Z3 F4WZC3 E1ZUW0 E2CA01 A0A1B6EG99 A0A087ZWJ7 A0A0B5ELM2 A0A2A3EMD0 A0A026WQ51 A0A0L7QVQ2 A0A310SL36 A0A195AUQ7 A0A158NHL6 E0VI01 A0A0C9RTP2 A0A195C1G5 A0A195ETP7 K7IUB7 U4UP84 A0A1S4E906 A0A1S4E9E2 N6T239 A0A182XCH8 A0A182WHI9 A0A182V6B5 A0A182MB94 A0A182JM09 Q7PM11 A0A182F1L7 A0A1S4H1I7 A0A182L5Z2 A0A182ICM5 A0A182UFE1 A0A182K756 A0A182RRI1 A7LBJ7 A0A182Q9P3 W5JQN6 A0A182Y5L4 A0A182GZ58 A0A182NUX2 Q16WY0 B0WF08 W8BII6 A0A182P3C8 A0A1I8PM32 A0A1I8PLZ9 A0A1B6HNM4 A0A0M4E690 B4NY93 A0A0R1DN47 B3N605 A0A0Q5WA44 B4KKY4 A8DZ06 X2J9Z3 A0A0J9TNV7 A0A0J9R2T0 W8BVD8 A0A0L0CMN0 W8AW12 Q29KE9 A0A0R3NUP1 A0A1I8NBR2 A0A3B0JNG7 B4N016 A0A336LYX8 A0A1W4V2R8 A0A1W4VGC6 A0A0Q9VZ03 A0A336KCU2 A0A1B0FKN7 B3ML95 A0A0P8XE51 A0A1A9X488 A0A154P2S8 B4HXS2 A0A1J1IL18 B4Q6G9 B4HBY5 B4JQ69 A0A2S2R5B3 A0A2H8TFU7 J9K4F6 D0IQF5
A0A067RSJ0 A0A1W4WYM4 D7EJQ5 A0A139WP66 A0A1W4WNW6 A0A1Y1NDS7 A0A151IZ18 A0A0N0BJP4 A0A3L8D6Z3 F4WZC3 E1ZUW0 E2CA01 A0A1B6EG99 A0A087ZWJ7 A0A0B5ELM2 A0A2A3EMD0 A0A026WQ51 A0A0L7QVQ2 A0A310SL36 A0A195AUQ7 A0A158NHL6 E0VI01 A0A0C9RTP2 A0A195C1G5 A0A195ETP7 K7IUB7 U4UP84 A0A1S4E906 A0A1S4E9E2 N6T239 A0A182XCH8 A0A182WHI9 A0A182V6B5 A0A182MB94 A0A182JM09 Q7PM11 A0A182F1L7 A0A1S4H1I7 A0A182L5Z2 A0A182ICM5 A0A182UFE1 A0A182K756 A0A182RRI1 A7LBJ7 A0A182Q9P3 W5JQN6 A0A182Y5L4 A0A182GZ58 A0A182NUX2 Q16WY0 B0WF08 W8BII6 A0A182P3C8 A0A1I8PM32 A0A1I8PLZ9 A0A1B6HNM4 A0A0M4E690 B4NY93 A0A0R1DN47 B3N605 A0A0Q5WA44 B4KKY4 A8DZ06 X2J9Z3 A0A0J9TNV7 A0A0J9R2T0 W8BVD8 A0A0L0CMN0 W8AW12 Q29KE9 A0A0R3NUP1 A0A1I8NBR2 A0A3B0JNG7 B4N016 A0A336LYX8 A0A1W4V2R8 A0A1W4VGC6 A0A0Q9VZ03 A0A336KCU2 A0A1B0FKN7 B3ML95 A0A0P8XE51 A0A1A9X488 A0A154P2S8 B4HXS2 A0A1J1IL18 B4Q6G9 B4HBY5 B4JQ69 A0A2S2R5B3 A0A2H8TFU7 J9K4F6 D0IQF5
Pubmed
19121390
22118469
28756777
26354079
29403074
24845553
+ More
18362917 19820115 28004739 30249741 21719571 20798317 24508170 21347285 20566863 20075255 23537049 12364791 20966253 20920257 23761445 25244985 26483478 17510324 24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 26108605 15632085 25315136
18362917 19820115 28004739 30249741 21719571 20798317 24508170 21347285 20566863 20075255 23537049 12364791 20966253 20920257 23761445 25244985 26483478 17510324 24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 26108605 15632085 25315136
EMBL
BABH01007608
BABH01007609
BABH01007610
BABH01007611
AGBW02014037
OWR42366.1
+ More
KZ149960 PZC76366.1 KQ459597 KPI95026.1 KQ460226 KPJ16431.1 PYGN01002430 PSN30498.1 KK852486 KDR22779.1 KQ971307 EFA12820.2 KYB29565.1 GEZM01005528 JAV96114.1 KQ980745 KYN13652.1 KQ435713 KOX79241.1 QOIP01000012 RLU16039.1 GL888471 EGI60451.1 GL434297 EFN75052.1 GL453916 EFN75238.1 GEDC01000340 JAS36958.1 KJ485709 AJE70861.1 KZ288215 PBC32432.1 KK107131 EZA58162.1 KQ414727 KOC62611.1 KQ763441 OAD55095.1 KQ976737 KYM75973.1 ADTU01015941 ADTU01015942 ADTU01015943 ADTU01015944 ADTU01015945 ADTU01015946 ADTU01015947 ADTU01015948 ADTU01015949 ADTU01015950 DS235174 EEB13007.1 GBYB01012115 JAG81882.1 KQ978350 KYM94719.1 KQ981975 KYN31613.1 AAZX01005288 KB632384 ERL94313.1 APGK01047174 APGK01047175 KB741077 ENN74179.1 AXCM01001522 AAAB01008980 EAA14534.4 APCN01005734 EF990671 ABS30732.1 AXCN02000750 ADMH02000615 ETN65618.1 JXUM01098836 JXUM01098837 JXUM01098838 KQ564450 KXJ72255.1 CH477551 EAT39108.2 DS231913 EDS25916.1 GAMC01013474 JAB93081.1 GECU01031420 JAS76286.1 CP012523 ALC39821.1 CM000157 EDW89729.2 KRJ98676.1 CH954177 EDV58043.2 KQS70228.1 CH933807 EDW11714.2 AE014134 ABV53683.3 ABV53684.2 AGB93001.2 AGB93002.2 AHN54477.1 AHN54478.1 AAF53476.4 AHN54476.1 CM002910 KMY90395.1 KMY90396.1 GAMC01013476 JAB93079.1 JRES01000175 KNC33608.1 GAMC01013475 JAB93080.1 CH379061 EAL33226.3 KRT04823.1 OUUW01000006 SPP82423.1 CH963920 EDW77951.2 UFQS01000219 UFQT01000219 SSX01549.1 SSX21929.1 CH940654 KRF77992.1 SSX01548.1 SSX21928.1 CCAG010005032 CH902620 EDV30684.2 KPU72989.1 KQ434792 KZC05430.1 CH480818 EDW51852.1 CVRI01000054 CRL00856.1 CM000361 EDX05151.1 CH479272 EDW39996.1 CH916372 EDV99049.1 GGMS01015982 MBY85185.1 GFXV01001115 MBW12920.1 ABLF02036139 ABLF02036143 ABLF02036144 ABLF02036153 BT100193 ACY40027.1
KZ149960 PZC76366.1 KQ459597 KPI95026.1 KQ460226 KPJ16431.1 PYGN01002430 PSN30498.1 KK852486 KDR22779.1 KQ971307 EFA12820.2 KYB29565.1 GEZM01005528 JAV96114.1 KQ980745 KYN13652.1 KQ435713 KOX79241.1 QOIP01000012 RLU16039.1 GL888471 EGI60451.1 GL434297 EFN75052.1 GL453916 EFN75238.1 GEDC01000340 JAS36958.1 KJ485709 AJE70861.1 KZ288215 PBC32432.1 KK107131 EZA58162.1 KQ414727 KOC62611.1 KQ763441 OAD55095.1 KQ976737 KYM75973.1 ADTU01015941 ADTU01015942 ADTU01015943 ADTU01015944 ADTU01015945 ADTU01015946 ADTU01015947 ADTU01015948 ADTU01015949 ADTU01015950 DS235174 EEB13007.1 GBYB01012115 JAG81882.1 KQ978350 KYM94719.1 KQ981975 KYN31613.1 AAZX01005288 KB632384 ERL94313.1 APGK01047174 APGK01047175 KB741077 ENN74179.1 AXCM01001522 AAAB01008980 EAA14534.4 APCN01005734 EF990671 ABS30732.1 AXCN02000750 ADMH02000615 ETN65618.1 JXUM01098836 JXUM01098837 JXUM01098838 KQ564450 KXJ72255.1 CH477551 EAT39108.2 DS231913 EDS25916.1 GAMC01013474 JAB93081.1 GECU01031420 JAS76286.1 CP012523 ALC39821.1 CM000157 EDW89729.2 KRJ98676.1 CH954177 EDV58043.2 KQS70228.1 CH933807 EDW11714.2 AE014134 ABV53683.3 ABV53684.2 AGB93001.2 AGB93002.2 AHN54477.1 AHN54478.1 AAF53476.4 AHN54476.1 CM002910 KMY90395.1 KMY90396.1 GAMC01013476 JAB93079.1 JRES01000175 KNC33608.1 GAMC01013475 JAB93080.1 CH379061 EAL33226.3 KRT04823.1 OUUW01000006 SPP82423.1 CH963920 EDW77951.2 UFQS01000219 UFQT01000219 SSX01549.1 SSX21929.1 CH940654 KRF77992.1 SSX01548.1 SSX21928.1 CCAG010005032 CH902620 EDV30684.2 KPU72989.1 KQ434792 KZC05430.1 CH480818 EDW51852.1 CVRI01000054 CRL00856.1 CM000361 EDX05151.1 CH479272 EDW39996.1 CH916372 EDV99049.1 GGMS01015982 MBY85185.1 GFXV01001115 MBW12920.1 ABLF02036139 ABLF02036143 ABLF02036144 ABLF02036153 BT100193 ACY40027.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000245037
UP000027135
+ More
UP000192223 UP000007266 UP000078492 UP000053105 UP000279307 UP000007755 UP000000311 UP000008237 UP000005203 UP000242457 UP000053097 UP000053825 UP000078540 UP000005205 UP000009046 UP000078542 UP000078541 UP000002358 UP000030742 UP000079169 UP000019118 UP000076407 UP000075920 UP000075903 UP000075883 UP000075880 UP000007062 UP000069272 UP000075882 UP000075840 UP000075902 UP000075881 UP000075900 UP000075886 UP000000673 UP000076408 UP000069940 UP000249989 UP000075884 UP000008820 UP000002320 UP000075885 UP000095300 UP000092553 UP000002282 UP000008711 UP000009192 UP000000803 UP000037069 UP000001819 UP000095301 UP000268350 UP000007798 UP000192221 UP000008792 UP000092444 UP000007801 UP000091820 UP000076502 UP000001292 UP000183832 UP000000304 UP000008744 UP000001070 UP000007819
UP000192223 UP000007266 UP000078492 UP000053105 UP000279307 UP000007755 UP000000311 UP000008237 UP000005203 UP000242457 UP000053097 UP000053825 UP000078540 UP000005205 UP000009046 UP000078542 UP000078541 UP000002358 UP000030742 UP000079169 UP000019118 UP000076407 UP000075920 UP000075903 UP000075883 UP000075880 UP000007062 UP000069272 UP000075882 UP000075840 UP000075902 UP000075881 UP000075900 UP000075886 UP000000673 UP000076408 UP000069940 UP000249989 UP000075884 UP000008820 UP000002320 UP000075885 UP000095300 UP000092553 UP000002282 UP000008711 UP000009192 UP000000803 UP000037069 UP000001819 UP000095301 UP000268350 UP000007798 UP000192221 UP000008792 UP000092444 UP000007801 UP000091820 UP000076502 UP000001292 UP000183832 UP000000304 UP000008744 UP000001070 UP000007819
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J2B3
A0A212ELK1
A0A2W1BUN7
A0A194PNL7
A0A0N0PDD7
A0A2P8XES1
+ More
A0A067RSJ0 A0A1W4WYM4 D7EJQ5 A0A139WP66 A0A1W4WNW6 A0A1Y1NDS7 A0A151IZ18 A0A0N0BJP4 A0A3L8D6Z3 F4WZC3 E1ZUW0 E2CA01 A0A1B6EG99 A0A087ZWJ7 A0A0B5ELM2 A0A2A3EMD0 A0A026WQ51 A0A0L7QVQ2 A0A310SL36 A0A195AUQ7 A0A158NHL6 E0VI01 A0A0C9RTP2 A0A195C1G5 A0A195ETP7 K7IUB7 U4UP84 A0A1S4E906 A0A1S4E9E2 N6T239 A0A182XCH8 A0A182WHI9 A0A182V6B5 A0A182MB94 A0A182JM09 Q7PM11 A0A182F1L7 A0A1S4H1I7 A0A182L5Z2 A0A182ICM5 A0A182UFE1 A0A182K756 A0A182RRI1 A7LBJ7 A0A182Q9P3 W5JQN6 A0A182Y5L4 A0A182GZ58 A0A182NUX2 Q16WY0 B0WF08 W8BII6 A0A182P3C8 A0A1I8PM32 A0A1I8PLZ9 A0A1B6HNM4 A0A0M4E690 B4NY93 A0A0R1DN47 B3N605 A0A0Q5WA44 B4KKY4 A8DZ06 X2J9Z3 A0A0J9TNV7 A0A0J9R2T0 W8BVD8 A0A0L0CMN0 W8AW12 Q29KE9 A0A0R3NUP1 A0A1I8NBR2 A0A3B0JNG7 B4N016 A0A336LYX8 A0A1W4V2R8 A0A1W4VGC6 A0A0Q9VZ03 A0A336KCU2 A0A1B0FKN7 B3ML95 A0A0P8XE51 A0A1A9X488 A0A154P2S8 B4HXS2 A0A1J1IL18 B4Q6G9 B4HBY5 B4JQ69 A0A2S2R5B3 A0A2H8TFU7 J9K4F6 D0IQF5
A0A067RSJ0 A0A1W4WYM4 D7EJQ5 A0A139WP66 A0A1W4WNW6 A0A1Y1NDS7 A0A151IZ18 A0A0N0BJP4 A0A3L8D6Z3 F4WZC3 E1ZUW0 E2CA01 A0A1B6EG99 A0A087ZWJ7 A0A0B5ELM2 A0A2A3EMD0 A0A026WQ51 A0A0L7QVQ2 A0A310SL36 A0A195AUQ7 A0A158NHL6 E0VI01 A0A0C9RTP2 A0A195C1G5 A0A195ETP7 K7IUB7 U4UP84 A0A1S4E906 A0A1S4E9E2 N6T239 A0A182XCH8 A0A182WHI9 A0A182V6B5 A0A182MB94 A0A182JM09 Q7PM11 A0A182F1L7 A0A1S4H1I7 A0A182L5Z2 A0A182ICM5 A0A182UFE1 A0A182K756 A0A182RRI1 A7LBJ7 A0A182Q9P3 W5JQN6 A0A182Y5L4 A0A182GZ58 A0A182NUX2 Q16WY0 B0WF08 W8BII6 A0A182P3C8 A0A1I8PM32 A0A1I8PLZ9 A0A1B6HNM4 A0A0M4E690 B4NY93 A0A0R1DN47 B3N605 A0A0Q5WA44 B4KKY4 A8DZ06 X2J9Z3 A0A0J9TNV7 A0A0J9R2T0 W8BVD8 A0A0L0CMN0 W8AW12 Q29KE9 A0A0R3NUP1 A0A1I8NBR2 A0A3B0JNG7 B4N016 A0A336LYX8 A0A1W4V2R8 A0A1W4VGC6 A0A0Q9VZ03 A0A336KCU2 A0A1B0FKN7 B3ML95 A0A0P8XE51 A0A1A9X488 A0A154P2S8 B4HXS2 A0A1J1IL18 B4Q6G9 B4HBY5 B4JQ69 A0A2S2R5B3 A0A2H8TFU7 J9K4F6 D0IQF5
PDB
3JBR
E-value=6.38826e-44,
Score=448
Ontologies
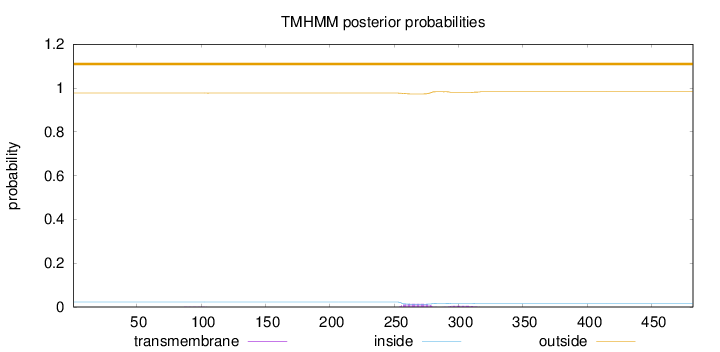
Topology
Length:
482
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.413910000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02311
outside
1 - 482
Population Genetic Test Statistics
Pi
201.854864
Theta
154.385774
Tajima's D
0.634527
CLR
0.628167
CSRT
0.554322283885806
Interpretation
Uncertain
