Gene
KWMTBOMO02896
Pre Gene Modal
BGIBMGA003650
Annotation
PREDICTED:_voltage-dependent_calcium_channel_subunit_alpha-2/delta-3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.405
Sequence
CDS
ATGGATTCCGATGAAAACTCCAGAGTCAACCTAACATCGCTACTTCTTGATCTGAGACACGATATGATCGAGCAGCGTGAAGGAGAAACCGAAATAGGTGTTAAAGTCCAATATGACAACATGAGGCGCGTGGCCACTAGGCGCCAGCGATACTTCTATAGCCCAGTCGACGGCACGCCTTTCTCTCTAGCCGCCGTGCTCCCTGACGGATACGGCATGTATGAGCTACAAGCTGAACAGGAAGTCAAACATAGCCCTATCAACGTAACGGAATATTTCAAAGGCGAAAACTGGAAGATTCATCCAGATTGGGTGTACTGCGAATACGCATCAACGTCAGACCAGACTTTCAACACGCCTGAAGAGCAGTTGCTGCATTTCTTGATCCGCGCCGGGAAACCTGGCTGGAAGTGGATGTCGTTGCGACCGAGAGCCCTGACGCTGCACAACGCGCACAAGAAACAGGATCGCGATTCTTATTACTGTGATAAAACTTTAGTTCAGTCGCTTATCAGGGATGCAATGGTAATTAAAGAATTGGACGCTCACACAGGACATGCCAATCACCACTCCCAACAAGGTCGGTTCCACAAGTTCGTGGTGACGTTGTCCTTTATCGCGACACGGAGCGGCCTACTGAAGTGGACTGAGAACTTCAATGGATCTCGGAATTCCGATTCTGCTGAACAGCATTTCAGCGAAAAGTACTCTCGTGCATTCGATTCAGAATGGTACCGTCGAGCCGTAGAGCATCACGCCATAGAGCCCGAGAGTTTCGTTTTCTCCGTTCCTTTCCGCGACGGATCCGAGGCCGAAGATCTGAGTGGAAGGCCTCCATTGGTGCTCGCCACTCACGCGGTGTTCGTCGAGTCGAGAGGTCACAGGGCGCCGGCGGCTGTCGTCGGTCTACACTTCCAACTTGACTCTCTCGCGAAACACTTTCTAAACGTAACATCAACGTGCACTGCTGGTACTGTTTGCAAGAAGACTTGTGCTGGTGACGAACTCGACTGTTATATTTTGGACGACAACGGATTTATCATTCTATCTGAAGACGTATCGCAGACAGGACTGTTCTTCGGCCAAGTCGACGGAACAATCATGGATTCACTAGTCCAAGATCGGATTTACAAGAAGGTCACTGTTCACGACCATCAAGGAAGGTGTCCCGATTCTAGAAATCCATTCAGCGGGAATTCTAATAAACTATCGCCTTTAAAACCACTTTCTTGGCTCGGTGGCTATCTAACTAATTTGTTGGCTATTTGGTTCCCGATGTGGACTCATGCAAGCGCTCATACGGCTCCTTATAACGAGGAAGAATTAGATTACGAGGACTACGAAGGCGAAGATGTCGACCACGATGATGATACCGATCGCGATCGAGGAACATACGAGATCATCTTGGACGGAGGAGGAGAGGGTCCGCGTGGGGGACCGGCCGGCGCTCCTCCCCCACCGCCTCCCGCCCCCTCACCGCGGGAACCCTCCCGTGAAGCGGTGCGTTCTGCAGCCGTGCGACCGTGCGATACTAGCGCTGAACTCTTCACGTTACAGTCGACCAGACTCAGCGCTGCGCAGCCTTTGAAAGGGAAACTTACTAATTGCCATAATTCTGGATGCGAAAGACCGTTCAGCGTCCAGAAGATCCCGCACAGCAACCTGATACTGTTGGTGGTGGACACGCTGTGCCCGTGCGGCGCCAAGCGGCTGGACGTACGCGCACGCGAGTCAGCCCCGCGCCCCACGTGCTCCTCCCCCCCGCCGCGCCCCCACCGCCGCCGACCTCATACTTGCGTCTCCTACCATCCTGAGGTCAGATTTTAG
Protein
MDSDENSRVNLTSLLLDLRHDMIEQREGETEIGVKVQYDNMRRVATRRQRYFYSPVDGTPFSLAAVLPDGYGMYELQAEQEVKHSPINVTEYFKGENWKIHPDWVYCEYASTSDQTFNTPEEQLLHFLIRAGKPGWKWMSLRPRALTLHNAHKKQDRDSYYCDKTLVQSLIRDAMVIKELDAHTGHANHHSQQGRFHKFVVTLSFIATRSGLLKWTENFNGSRNSDSAEQHFSEKYSRAFDSEWYRRAVEHHAIEPESFVFSVPFRDGSEAEDLSGRPPLVLATHAVFVESRGHRAPAAVVGLHFQLDSLAKHFLNVTSTCTAGTVCKKTCAGDELDCYILDDNGFIILSEDVSQTGLFFGQVDGTIMDSLVQDRIYKKVTVHDHQGRCPDSRNPFSGNSNKLSPLKPLSWLGGYLTNLLAIWFPMWTHASAHTAPYNEEELDYEDYEGEDVDHDDDTDRDRGTYEIILDGGGEGPRGGPAGAPPPPPPAPSPREPSREAVRSAAVRPCDTSAELFTLQSTRLSAAQPLKGKLTNCHNSGCERPFSVQKIPHSNLILLVVDTLCPCGAKRLDVRARESAPRPTCSSPPPRPHRRRPHTCVSYHPEVRF
Summary
Uniprot
A0A2H1V9U9
A0A212ELK1
A0A2A4J1S3
A0A194PNL7
A0A0N0PDD7
H9J2B3
+ More
A0A0L7LD67 A0A182WHI9 A0A182MB94 A0A1W4WNW6 A0A1W4WYM4 A0A182UFE1 A0A182RRI1 A0A182P3C8 A0A182JM09 A0A182NUX2 A0A182V6B5 A0A182Q9P3 A0A1S4H1I7 A0A182L5Z2 A0A182ICM5 A0A1I8PM32 A0A182Y5L4 A0A182K756 A0A1A9Y575 A7LBJ7 B4JQ69 W8BVD8 A0A0Q9VZ03 A0A1I8PLZ9 A0A1I8NBR2 B4KKY4 A0A3L8D6Z3 B4N016 A0A182F1L7 B3ML95 A0A1B0BLF9 A0A0L0CMN0 A0A3B0JNG7 A0A1B0FKN7 W8AW12 Q29KE9 A0A1W4V2R8 A0A2H8TFU7 B4HBY5 A0A0P8XE51 A0A232FGE5 W5JQN6 A0A182GZ58 A0A1W4VGC6 A0A0R3NUP1 B3N605 A0A0J9TNV7 A8DZ06 Q16WY0 B4NY93 A0A1B0CA97 A0A0Q5WA44 J9K4F6 A0A0J9R2T0 X2J9Z3 A0A0R1DN47 A0A1A9X488 A0A1J1IL18 Q7PM11 B4Q6G9 B4HXS2 A0A0M4E690 U4UP84 N6T239 A0A1B0D6Z4 A0A0P5KXR2
A0A0L7LD67 A0A182WHI9 A0A182MB94 A0A1W4WNW6 A0A1W4WYM4 A0A182UFE1 A0A182RRI1 A0A182P3C8 A0A182JM09 A0A182NUX2 A0A182V6B5 A0A182Q9P3 A0A1S4H1I7 A0A182L5Z2 A0A182ICM5 A0A1I8PM32 A0A182Y5L4 A0A182K756 A0A1A9Y575 A7LBJ7 B4JQ69 W8BVD8 A0A0Q9VZ03 A0A1I8PLZ9 A0A1I8NBR2 B4KKY4 A0A3L8D6Z3 B4N016 A0A182F1L7 B3ML95 A0A1B0BLF9 A0A0L0CMN0 A0A3B0JNG7 A0A1B0FKN7 W8AW12 Q29KE9 A0A1W4V2R8 A0A2H8TFU7 B4HBY5 A0A0P8XE51 A0A232FGE5 W5JQN6 A0A182GZ58 A0A1W4VGC6 A0A0R3NUP1 B3N605 A0A0J9TNV7 A8DZ06 Q16WY0 B4NY93 A0A1B0CA97 A0A0Q5WA44 J9K4F6 A0A0J9R2T0 X2J9Z3 A0A0R1DN47 A0A1A9X488 A0A1J1IL18 Q7PM11 B4Q6G9 B4HXS2 A0A0M4E690 U4UP84 N6T239 A0A1B0D6Z4 A0A0P5KXR2
Pubmed
EMBL
ODYU01001433
SOQ37609.1
AGBW02014037
OWR42366.1
NWSH01003681
PCG65941.1
+ More
KQ459597 KPI95026.1 KQ460226 KPJ16431.1 BABH01007608 BABH01007609 BABH01007610 BABH01007611 JTDY01001694 KOB73141.1 AXCM01001522 AXCN02000750 AAAB01008980 APCN01005734 EF990671 ABS30732.1 CH916372 EDV99049.1 GAMC01013476 JAB93079.1 CH940654 KRF77992.1 CH933807 EDW11714.2 QOIP01000012 RLU16039.1 CH963920 EDW77951.2 CH902620 EDV30684.2 JXJN01016335 JRES01000175 KNC33608.1 OUUW01000006 SPP82423.1 CCAG010005032 GAMC01013475 JAB93080.1 CH379061 EAL33226.3 GFXV01001115 MBW12920.1 CH479272 EDW39996.1 KPU72989.1 NNAY01000284 OXU29499.1 ADMH02000615 ETN65618.1 JXUM01098836 JXUM01098837 JXUM01098838 KQ564450 KXJ72255.1 KRT04823.1 CH954177 EDV58043.2 CM002910 KMY90395.1 AE014134 ABV53683.3 ABV53684.2 AGB93001.2 AGB93002.2 AHN54477.1 AHN54478.1 CH477551 EAT39108.2 CM000157 EDW89729.2 AJWK01003448 AJWK01003449 AJWK01003450 AJWK01003451 AJWK01003452 AJWK01003453 KQS70228.1 ABLF02036139 ABLF02036143 ABLF02036144 ABLF02036153 KMY90396.1 AAF53476.4 AHN54476.1 KRJ98676.1 CVRI01000054 CRL00856.1 EAA14534.4 CM000361 EDX05151.1 CH480818 EDW51852.1 CP012523 ALC39821.1 KB632384 ERL94313.1 APGK01047174 APGK01047175 KB741077 ENN74179.1 AJVK01003831 AJVK01003832 AJVK01003833 GDIQ01203591 JAK48134.1
KQ459597 KPI95026.1 KQ460226 KPJ16431.1 BABH01007608 BABH01007609 BABH01007610 BABH01007611 JTDY01001694 KOB73141.1 AXCM01001522 AXCN02000750 AAAB01008980 APCN01005734 EF990671 ABS30732.1 CH916372 EDV99049.1 GAMC01013476 JAB93079.1 CH940654 KRF77992.1 CH933807 EDW11714.2 QOIP01000012 RLU16039.1 CH963920 EDW77951.2 CH902620 EDV30684.2 JXJN01016335 JRES01000175 KNC33608.1 OUUW01000006 SPP82423.1 CCAG010005032 GAMC01013475 JAB93080.1 CH379061 EAL33226.3 GFXV01001115 MBW12920.1 CH479272 EDW39996.1 KPU72989.1 NNAY01000284 OXU29499.1 ADMH02000615 ETN65618.1 JXUM01098836 JXUM01098837 JXUM01098838 KQ564450 KXJ72255.1 KRT04823.1 CH954177 EDV58043.2 CM002910 KMY90395.1 AE014134 ABV53683.3 ABV53684.2 AGB93001.2 AGB93002.2 AHN54477.1 AHN54478.1 CH477551 EAT39108.2 CM000157 EDW89729.2 AJWK01003448 AJWK01003449 AJWK01003450 AJWK01003451 AJWK01003452 AJWK01003453 KQS70228.1 ABLF02036139 ABLF02036143 ABLF02036144 ABLF02036153 KMY90396.1 AAF53476.4 AHN54476.1 KRJ98676.1 CVRI01000054 CRL00856.1 EAA14534.4 CM000361 EDX05151.1 CH480818 EDW51852.1 CP012523 ALC39821.1 KB632384 ERL94313.1 APGK01047174 APGK01047175 KB741077 ENN74179.1 AJVK01003831 AJVK01003832 AJVK01003833 GDIQ01203591 JAK48134.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000005204
UP000037510
+ More
UP000075920 UP000075883 UP000192223 UP000075902 UP000075900 UP000075885 UP000075880 UP000075884 UP000075903 UP000075886 UP000075882 UP000075840 UP000095300 UP000076408 UP000075881 UP000092443 UP000001070 UP000008792 UP000095301 UP000009192 UP000279307 UP000007798 UP000069272 UP000007801 UP000092460 UP000037069 UP000268350 UP000092444 UP000001819 UP000192221 UP000008744 UP000215335 UP000000673 UP000069940 UP000249989 UP000008711 UP000000803 UP000008820 UP000002282 UP000092461 UP000007819 UP000091820 UP000183832 UP000007062 UP000000304 UP000001292 UP000092553 UP000030742 UP000019118 UP000092462
UP000075920 UP000075883 UP000192223 UP000075902 UP000075900 UP000075885 UP000075880 UP000075884 UP000075903 UP000075886 UP000075882 UP000075840 UP000095300 UP000076408 UP000075881 UP000092443 UP000001070 UP000008792 UP000095301 UP000009192 UP000279307 UP000007798 UP000069272 UP000007801 UP000092460 UP000037069 UP000268350 UP000092444 UP000001819 UP000192221 UP000008744 UP000215335 UP000000673 UP000069940 UP000249989 UP000008711 UP000000803 UP000008820 UP000002282 UP000092461 UP000007819 UP000091820 UP000183832 UP000007062 UP000000304 UP000001292 UP000092553 UP000030742 UP000019118 UP000092462
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1V9U9
A0A212ELK1
A0A2A4J1S3
A0A194PNL7
A0A0N0PDD7
H9J2B3
+ More
A0A0L7LD67 A0A182WHI9 A0A182MB94 A0A1W4WNW6 A0A1W4WYM4 A0A182UFE1 A0A182RRI1 A0A182P3C8 A0A182JM09 A0A182NUX2 A0A182V6B5 A0A182Q9P3 A0A1S4H1I7 A0A182L5Z2 A0A182ICM5 A0A1I8PM32 A0A182Y5L4 A0A182K756 A0A1A9Y575 A7LBJ7 B4JQ69 W8BVD8 A0A0Q9VZ03 A0A1I8PLZ9 A0A1I8NBR2 B4KKY4 A0A3L8D6Z3 B4N016 A0A182F1L7 B3ML95 A0A1B0BLF9 A0A0L0CMN0 A0A3B0JNG7 A0A1B0FKN7 W8AW12 Q29KE9 A0A1W4V2R8 A0A2H8TFU7 B4HBY5 A0A0P8XE51 A0A232FGE5 W5JQN6 A0A182GZ58 A0A1W4VGC6 A0A0R3NUP1 B3N605 A0A0J9TNV7 A8DZ06 Q16WY0 B4NY93 A0A1B0CA97 A0A0Q5WA44 J9K4F6 A0A0J9R2T0 X2J9Z3 A0A0R1DN47 A0A1A9X488 A0A1J1IL18 Q7PM11 B4Q6G9 B4HXS2 A0A0M4E690 U4UP84 N6T239 A0A1B0D6Z4 A0A0P5KXR2
A0A0L7LD67 A0A182WHI9 A0A182MB94 A0A1W4WNW6 A0A1W4WYM4 A0A182UFE1 A0A182RRI1 A0A182P3C8 A0A182JM09 A0A182NUX2 A0A182V6B5 A0A182Q9P3 A0A1S4H1I7 A0A182L5Z2 A0A182ICM5 A0A1I8PM32 A0A182Y5L4 A0A182K756 A0A1A9Y575 A7LBJ7 B4JQ69 W8BVD8 A0A0Q9VZ03 A0A1I8PLZ9 A0A1I8NBR2 B4KKY4 A0A3L8D6Z3 B4N016 A0A182F1L7 B3ML95 A0A1B0BLF9 A0A0L0CMN0 A0A3B0JNG7 A0A1B0FKN7 W8AW12 Q29KE9 A0A1W4V2R8 A0A2H8TFU7 B4HBY5 A0A0P8XE51 A0A232FGE5 W5JQN6 A0A182GZ58 A0A1W4VGC6 A0A0R3NUP1 B3N605 A0A0J9TNV7 A8DZ06 Q16WY0 B4NY93 A0A1B0CA97 A0A0Q5WA44 J9K4F6 A0A0J9R2T0 X2J9Z3 A0A0R1DN47 A0A1A9X488 A0A1J1IL18 Q7PM11 B4Q6G9 B4HXS2 A0A0M4E690 U4UP84 N6T239 A0A1B0D6Z4 A0A0P5KXR2
PDB
5GJW
E-value=3.77054e-18,
Score=226
Ontologies
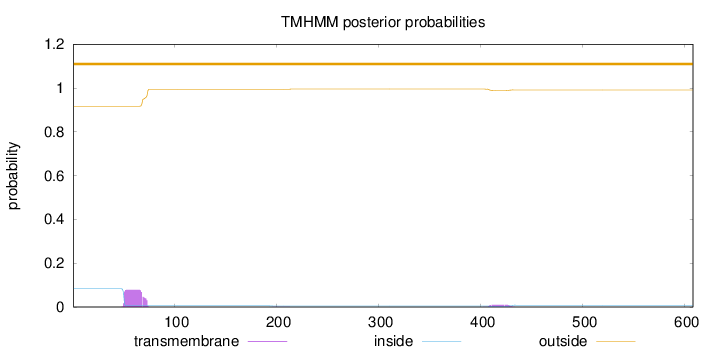
Topology
Length:
608
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.82137
Exp number, first 60 AAs:
0.79018
Total prob of N-in:
0.08318
outside
1 - 608
Population Genetic Test Statistics
Pi
173.233676
Theta
161.664375
Tajima's D
0.622201
CLR
0.191025
CSRT
0.542922853857307
Interpretation
Uncertain
