Gene
KWMTBOMO02893 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003652
Annotation
Dm0-like_lamin_[Spodoptera_frugiperda]
Location in the cell
Nuclear Reliability : 3.17
Sequence
CDS
ATGGCCGCCCAGCTGCGGGAGTACGCCACGCTCATGGACATCAAGATATCGCTCGACCGTGAAATCTCTACCTACAGAGCACTTCTTGAAGGAGAGGAAGACAGGTTAAACCTGACATCTCAGTCGCCGGGGCGAGAGTCTCGCGCGTCAGGCAGCGGCAGCGCGTCGGCCAGCGGGTCCCGCGTCACGCCGTCCCGCCGCGCCACGCCGCTGCGCGCCGCACGCAAGCGCACCCTGCTCGACGAGACCGAGGAGCGCTCGCTGCAGGACTTCAGCGTCACCTCCAGCGCCAAGGGAGACCTCGAGGTCGCCGAGGCCTGCCCCGATGGAACGTTCGTGAAGATCAGGAACAAGGGCAAGAAGGAATTAAGCCTCGGCGGATATCAGATCCTGCGCAAGGCGGGTGATCAAGAAACCGTGTTCAAATTCCATCGCACCGTGAAGCTGGAGCCGGGCGCCGTGAGCACGGTGTGGTCGGCCGACACGGGCGCCTCGCACGACCCGCCGCACGACATCGTCATGAAGGGACAGAAGTGGTTCGTGGCCGATACCTTCACCACGGCTCTGCTCAACAACGAACAGGAGGAAGTCGCAATTTCTGAACGTCAACGACGACAGATAAGCACGAGCGCACAGAGACACCGCGAGCTCGCACACAAGTATCCGCGCCGAGAACAGATCGGAGAGATCCGCGAAGGTGAAGAGAATTGCCGCATCATGTAA
Protein
MAAQLREYATLMDIKISLDREISTYRALLEGEEDRLNLTSQSPGRESRASGSGSASASGSRVTPSRRATPLRAARKRTLLDETEERSLQDFSVTSSAKGDLEVAEACPDGTFVKIRNKGKKELSLGGYQILRKAGDQETVFKFHRTVKLEPGAVSTVWSADTGASHDPPHDIVMKGQKWFVADTFTTALLNNEQEEVAISERQRRQISTSAQRHRELAHKYPRREQIGEIREGEENCRIM
Summary
Similarity
Belongs to the intermediate filament family.
Uniprot
H9J2B5
A0A2H1V9Y5
A0A0P0IVE9
S4PXW2
A0A2W1BMT6
A0A0N0PDR8
+ More
A0A194PNK2 A0A212ELM3 A0A1E1WWA4 A0A0L7L1G8 A0A0L7QWW3 K7ISC5 A0A154PEI5 A0A2A3EM64 A0A1Y1NFL4 A0A088AD32 A0A232F9J9 E9IKN7 E2AVV9 A0A026W841 A0A182K275 A0A0M8ZSD0 E2C4M7 A0A182TNQ5 A0A336LYN6 A0A182STQ7 A0A0C9R470 F4W5M2 A0A1B0CDW5 A0A0C9PKT8 A0A0C9R9V0 A0A182GN94 A0A182Y9I8 A0A158NSD7 A0A1Y1NC18 A0A182XJQ7 A0A182L578 A0A182HHM7 A0A182URP7 A0A336MTG3 V5GWB7 A0A182RXV6 Q7PZJ0 Q17FG6 A0A1B6DQH4 A0A182M7I4 A0A1B0DD59 A0A151WLV6 A0A151HYG0 D2A5Q1 A0A195ES10 A0A023ETW9 A0A1Q3F388 A0A182QBU3 T1DRP1 A0A182WE19 A0A182PMW4 A0A2J7Q3M4 B0WS03 A0A2M4BND1 A0A182FZW7 A0A2M4AE54 A0A1L8DX61 U5EN31 A0A0J7KKM5 A0A0T6ASS6 A0A2M3Z5K3 A0A182JFM2 A0A2M4BJ15 W5JU89 A0A182N2S1 A0A084WIH0 A0A0K8V002 T1GAR3 A0A034VUV3 A0A1W4WZ32 W8B4Q8 A0A0A1XLP3 A0A0N8CKM0 A0A0P6IZ96 N6U8V8 U4UCY7 A0A0P5A6Y2 A0A0P4YV03 A0A0P5FJ80 A0A0N8DYU1 A0A0P5RHG7 A0A0P6D4Z4 A0A0P5TX23 A0A0N8BYF3 A0A0P5V0Q0 A0A0P5TP46 A0A0P5TF75 A0A0P5VLS0 A0A0P5PEJ9 A0A0P5YUD9 A0A0P6HNL1 A0A067R8L2 A0A0P5NEP2 A0A0N8DP35
A0A194PNK2 A0A212ELM3 A0A1E1WWA4 A0A0L7L1G8 A0A0L7QWW3 K7ISC5 A0A154PEI5 A0A2A3EM64 A0A1Y1NFL4 A0A088AD32 A0A232F9J9 E9IKN7 E2AVV9 A0A026W841 A0A182K275 A0A0M8ZSD0 E2C4M7 A0A182TNQ5 A0A336LYN6 A0A182STQ7 A0A0C9R470 F4W5M2 A0A1B0CDW5 A0A0C9PKT8 A0A0C9R9V0 A0A182GN94 A0A182Y9I8 A0A158NSD7 A0A1Y1NC18 A0A182XJQ7 A0A182L578 A0A182HHM7 A0A182URP7 A0A336MTG3 V5GWB7 A0A182RXV6 Q7PZJ0 Q17FG6 A0A1B6DQH4 A0A182M7I4 A0A1B0DD59 A0A151WLV6 A0A151HYG0 D2A5Q1 A0A195ES10 A0A023ETW9 A0A1Q3F388 A0A182QBU3 T1DRP1 A0A182WE19 A0A182PMW4 A0A2J7Q3M4 B0WS03 A0A2M4BND1 A0A182FZW7 A0A2M4AE54 A0A1L8DX61 U5EN31 A0A0J7KKM5 A0A0T6ASS6 A0A2M3Z5K3 A0A182JFM2 A0A2M4BJ15 W5JU89 A0A182N2S1 A0A084WIH0 A0A0K8V002 T1GAR3 A0A034VUV3 A0A1W4WZ32 W8B4Q8 A0A0A1XLP3 A0A0N8CKM0 A0A0P6IZ96 N6U8V8 U4UCY7 A0A0P5A6Y2 A0A0P4YV03 A0A0P5FJ80 A0A0N8DYU1 A0A0P5RHG7 A0A0P6D4Z4 A0A0P5TX23 A0A0N8BYF3 A0A0P5V0Q0 A0A0P5TP46 A0A0P5TF75 A0A0P5VLS0 A0A0P5PEJ9 A0A0P5YUD9 A0A0P6HNL1 A0A067R8L2 A0A0P5NEP2 A0A0N8DP35
Pubmed
EMBL
BABH01007612
ODYU01001433
SOQ37611.1
KT318393
ALK01985.1
GAIX01004343
+ More
JAA88217.1 KZ149960 PZC76369.1 KQ460226 KPJ16419.1 KQ459597 KPI95011.1 AGBW02014037 OWR42369.1 GDQN01000003 JAT91051.1 JTDY01003712 KOB69109.1 KQ414706 KOC63056.1 KQ434889 KZC10286.1 KZ288220 PBC32131.1 GEZM01007082 GEZM01007081 JAV95415.1 NNAY01000590 OXU27516.1 GL764021 EFZ18875.1 GL443213 EFN62381.1 KK107347 QOIP01000007 EZA52277.1 RLU20880.1 KQ435878 KOX69977.1 GL452536 EFN77107.1 UFQS01000212 UFQT01000212 SSX01393.1 SSX21773.1 GBYB01001616 JAG71383.1 GL887655 EGI70519.1 AJWK01008307 AJWK01008308 AJWK01008309 AJWK01008310 AJWK01008311 GBYB01001618 JAG71385.1 GBYB01009717 JAG79484.1 JXUM01076115 JXUM01076116 KQ562925 KXJ74823.1 ADTU01024851 ADTU01024852 ADTU01024853 GEZM01007083 JAV95413.1 APCN01002429 UFQT01001875 SSX32063.1 GALX01003938 JAB64528.1 AAAB01008986 EAA00263.4 CH477271 EAT45279.1 GEDC01009380 JAS27918.1 AXCM01001841 AJVK01031668 AJVK01031669 KQ982956 KYQ48797.1 KQ976730 KYM76484.1 KQ971345 EFA05043.1 KQ981993 KYN31040.1 GAPW01000778 JAC12820.1 GFDL01013056 JAV21989.1 AXCN02001087 GAMD01000565 JAB01026.1 NEVH01019066 PNF23170.1 DS232063 EDS33621.1 GGFJ01005446 MBW54587.1 GGFK01005746 MBW39067.1 GFDF01003034 JAV11050.1 GANO01000751 JAB59120.1 LBMM01006237 KMQ90784.1 LJIG01022884 KRT78232.1 GGFM01003042 MBW23793.1 GGFJ01003918 MBW53059.1 ADMH02000120 ETN67706.1 ATLV01023931 KE525347 KFB50014.1 GDHF01020087 GDHF01011428 GDHF01010988 JAI32227.1 JAI40886.1 JAI41326.1 CAQQ02158731 GAKP01013065 JAC45887.1 GAMC01018269 JAB88286.1 GBXI01017213 GBXI01005197 GBXI01002789 JAC97078.1 JAD09095.1 JAD11503.1 GDIP01122487 JAL81227.1 GDIQ01002179 JAN92558.1 APGK01044203 APGK01044204 KB741021 ENN75032.1 KB632003 ERL87810.1 GDIP01207620 JAJ15782.1 GDIP01226875 JAI96526.1 GDIQ01254440 JAJ97284.1 GDIQ01078540 JAN16197.1 GDIQ01110024 GDIP01130334 JAL41702.1 JAL73380.1 GDIP01155389 GDIQ01111839 GDIQ01084945 LRGB01000248 JAJ68013.1 JAN09792.1 KZS20054.1 GDIP01120782 JAL82932.1 GDIQ01132635 JAL19091.1 GDIP01122486 GDIQ01060806 JAL81228.1 JAN33931.1 GDIP01124850 JAL78864.1 GDIP01127289 JAL76425.1 GDIP01098289 JAM05426.1 GDIQ01139428 JAL12298.1 GDIP01054556 JAM49159.1 GDIQ01016656 JAN78081.1 KK852632 KDR19813.1 GDIQ01143168 JAL08558.1 GDIP01014767 JAM88948.1
JAA88217.1 KZ149960 PZC76369.1 KQ460226 KPJ16419.1 KQ459597 KPI95011.1 AGBW02014037 OWR42369.1 GDQN01000003 JAT91051.1 JTDY01003712 KOB69109.1 KQ414706 KOC63056.1 KQ434889 KZC10286.1 KZ288220 PBC32131.1 GEZM01007082 GEZM01007081 JAV95415.1 NNAY01000590 OXU27516.1 GL764021 EFZ18875.1 GL443213 EFN62381.1 KK107347 QOIP01000007 EZA52277.1 RLU20880.1 KQ435878 KOX69977.1 GL452536 EFN77107.1 UFQS01000212 UFQT01000212 SSX01393.1 SSX21773.1 GBYB01001616 JAG71383.1 GL887655 EGI70519.1 AJWK01008307 AJWK01008308 AJWK01008309 AJWK01008310 AJWK01008311 GBYB01001618 JAG71385.1 GBYB01009717 JAG79484.1 JXUM01076115 JXUM01076116 KQ562925 KXJ74823.1 ADTU01024851 ADTU01024852 ADTU01024853 GEZM01007083 JAV95413.1 APCN01002429 UFQT01001875 SSX32063.1 GALX01003938 JAB64528.1 AAAB01008986 EAA00263.4 CH477271 EAT45279.1 GEDC01009380 JAS27918.1 AXCM01001841 AJVK01031668 AJVK01031669 KQ982956 KYQ48797.1 KQ976730 KYM76484.1 KQ971345 EFA05043.1 KQ981993 KYN31040.1 GAPW01000778 JAC12820.1 GFDL01013056 JAV21989.1 AXCN02001087 GAMD01000565 JAB01026.1 NEVH01019066 PNF23170.1 DS232063 EDS33621.1 GGFJ01005446 MBW54587.1 GGFK01005746 MBW39067.1 GFDF01003034 JAV11050.1 GANO01000751 JAB59120.1 LBMM01006237 KMQ90784.1 LJIG01022884 KRT78232.1 GGFM01003042 MBW23793.1 GGFJ01003918 MBW53059.1 ADMH02000120 ETN67706.1 ATLV01023931 KE525347 KFB50014.1 GDHF01020087 GDHF01011428 GDHF01010988 JAI32227.1 JAI40886.1 JAI41326.1 CAQQ02158731 GAKP01013065 JAC45887.1 GAMC01018269 JAB88286.1 GBXI01017213 GBXI01005197 GBXI01002789 JAC97078.1 JAD09095.1 JAD11503.1 GDIP01122487 JAL81227.1 GDIQ01002179 JAN92558.1 APGK01044203 APGK01044204 KB741021 ENN75032.1 KB632003 ERL87810.1 GDIP01207620 JAJ15782.1 GDIP01226875 JAI96526.1 GDIQ01254440 JAJ97284.1 GDIQ01078540 JAN16197.1 GDIQ01110024 GDIP01130334 JAL41702.1 JAL73380.1 GDIP01155389 GDIQ01111839 GDIQ01084945 LRGB01000248 JAJ68013.1 JAN09792.1 KZS20054.1 GDIP01120782 JAL82932.1 GDIQ01132635 JAL19091.1 GDIP01122486 GDIQ01060806 JAL81228.1 JAN33931.1 GDIP01124850 JAL78864.1 GDIP01127289 JAL76425.1 GDIP01098289 JAM05426.1 GDIQ01139428 JAL12298.1 GDIP01054556 JAM49159.1 GDIQ01016656 JAN78081.1 KK852632 KDR19813.1 GDIQ01143168 JAL08558.1 GDIP01014767 JAM88948.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000053825
+ More
UP000002358 UP000076502 UP000242457 UP000005203 UP000215335 UP000000311 UP000053097 UP000279307 UP000075881 UP000053105 UP000008237 UP000075902 UP000075901 UP000007755 UP000092461 UP000069940 UP000249989 UP000076408 UP000005205 UP000076407 UP000075882 UP000075840 UP000075903 UP000075900 UP000007062 UP000008820 UP000075883 UP000092462 UP000075809 UP000078540 UP000007266 UP000078541 UP000075886 UP000075920 UP000075885 UP000235965 UP000002320 UP000069272 UP000036403 UP000075880 UP000000673 UP000075884 UP000030765 UP000015102 UP000192223 UP000019118 UP000030742 UP000076858 UP000027135
UP000002358 UP000076502 UP000242457 UP000005203 UP000215335 UP000000311 UP000053097 UP000279307 UP000075881 UP000053105 UP000008237 UP000075902 UP000075901 UP000007755 UP000092461 UP000069940 UP000249989 UP000076408 UP000005205 UP000076407 UP000075882 UP000075840 UP000075903 UP000075900 UP000007062 UP000008820 UP000075883 UP000092462 UP000075809 UP000078540 UP000007266 UP000078541 UP000075886 UP000075920 UP000075885 UP000235965 UP000002320 UP000069272 UP000036403 UP000075880 UP000000673 UP000075884 UP000030765 UP000015102 UP000192223 UP000019118 UP000030742 UP000076858 UP000027135
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J2B5
A0A2H1V9Y5
A0A0P0IVE9
S4PXW2
A0A2W1BMT6
A0A0N0PDR8
+ More
A0A194PNK2 A0A212ELM3 A0A1E1WWA4 A0A0L7L1G8 A0A0L7QWW3 K7ISC5 A0A154PEI5 A0A2A3EM64 A0A1Y1NFL4 A0A088AD32 A0A232F9J9 E9IKN7 E2AVV9 A0A026W841 A0A182K275 A0A0M8ZSD0 E2C4M7 A0A182TNQ5 A0A336LYN6 A0A182STQ7 A0A0C9R470 F4W5M2 A0A1B0CDW5 A0A0C9PKT8 A0A0C9R9V0 A0A182GN94 A0A182Y9I8 A0A158NSD7 A0A1Y1NC18 A0A182XJQ7 A0A182L578 A0A182HHM7 A0A182URP7 A0A336MTG3 V5GWB7 A0A182RXV6 Q7PZJ0 Q17FG6 A0A1B6DQH4 A0A182M7I4 A0A1B0DD59 A0A151WLV6 A0A151HYG0 D2A5Q1 A0A195ES10 A0A023ETW9 A0A1Q3F388 A0A182QBU3 T1DRP1 A0A182WE19 A0A182PMW4 A0A2J7Q3M4 B0WS03 A0A2M4BND1 A0A182FZW7 A0A2M4AE54 A0A1L8DX61 U5EN31 A0A0J7KKM5 A0A0T6ASS6 A0A2M3Z5K3 A0A182JFM2 A0A2M4BJ15 W5JU89 A0A182N2S1 A0A084WIH0 A0A0K8V002 T1GAR3 A0A034VUV3 A0A1W4WZ32 W8B4Q8 A0A0A1XLP3 A0A0N8CKM0 A0A0P6IZ96 N6U8V8 U4UCY7 A0A0P5A6Y2 A0A0P4YV03 A0A0P5FJ80 A0A0N8DYU1 A0A0P5RHG7 A0A0P6D4Z4 A0A0P5TX23 A0A0N8BYF3 A0A0P5V0Q0 A0A0P5TP46 A0A0P5TF75 A0A0P5VLS0 A0A0P5PEJ9 A0A0P5YUD9 A0A0P6HNL1 A0A067R8L2 A0A0P5NEP2 A0A0N8DP35
A0A194PNK2 A0A212ELM3 A0A1E1WWA4 A0A0L7L1G8 A0A0L7QWW3 K7ISC5 A0A154PEI5 A0A2A3EM64 A0A1Y1NFL4 A0A088AD32 A0A232F9J9 E9IKN7 E2AVV9 A0A026W841 A0A182K275 A0A0M8ZSD0 E2C4M7 A0A182TNQ5 A0A336LYN6 A0A182STQ7 A0A0C9R470 F4W5M2 A0A1B0CDW5 A0A0C9PKT8 A0A0C9R9V0 A0A182GN94 A0A182Y9I8 A0A158NSD7 A0A1Y1NC18 A0A182XJQ7 A0A182L578 A0A182HHM7 A0A182URP7 A0A336MTG3 V5GWB7 A0A182RXV6 Q7PZJ0 Q17FG6 A0A1B6DQH4 A0A182M7I4 A0A1B0DD59 A0A151WLV6 A0A151HYG0 D2A5Q1 A0A195ES10 A0A023ETW9 A0A1Q3F388 A0A182QBU3 T1DRP1 A0A182WE19 A0A182PMW4 A0A2J7Q3M4 B0WS03 A0A2M4BND1 A0A182FZW7 A0A2M4AE54 A0A1L8DX61 U5EN31 A0A0J7KKM5 A0A0T6ASS6 A0A2M3Z5K3 A0A182JFM2 A0A2M4BJ15 W5JU89 A0A182N2S1 A0A084WIH0 A0A0K8V002 T1GAR3 A0A034VUV3 A0A1W4WZ32 W8B4Q8 A0A0A1XLP3 A0A0N8CKM0 A0A0P6IZ96 N6U8V8 U4UCY7 A0A0P5A6Y2 A0A0P4YV03 A0A0P5FJ80 A0A0N8DYU1 A0A0P5RHG7 A0A0P6D4Z4 A0A0P5TX23 A0A0N8BYF3 A0A0P5V0Q0 A0A0P5TP46 A0A0P5TF75 A0A0P5VLS0 A0A0P5PEJ9 A0A0P5YUD9 A0A0P6HNL1 A0A067R8L2 A0A0P5NEP2 A0A0N8DP35
PDB
1IVT
E-value=4.46429e-15,
Score=195
Ontologies
GO
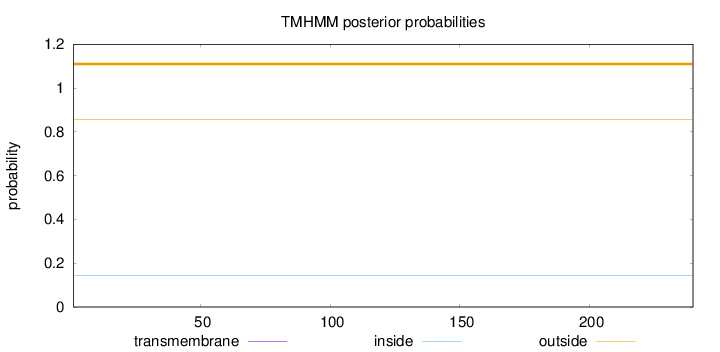
Topology
Length:
240
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14248
outside
1 - 240
Population Genetic Test Statistics
Pi
234.194491
Theta
217.105697
Tajima's D
0.533977
CLR
0.966251
CSRT
0.524323783810809
Interpretation
Uncertain
