Gene
KWMTBOMO02891
Pre Gene Modal
BGIBMGA003654
Annotation
PREDICTED:_insulin_receptor_isoform_X2_[Bombyx_mori]
Full name
Tyrosine-protein kinase receptor
+ More
Insulin-like receptor
Insulin-like receptor
Location in the cell
Extracellular Reliability : 2.361
Sequence
CDS
ATGGACCTACGAAACAATCCGGACCAGATCAGACGTTTGGAAGGATGTCGGGTGATCGAGGGTCAGCTCAGTATCGTTCTGATGGAGCGGGCCACAAATCAAATATTCGACAACATGAGCTTCCCGCAGTTAAGGGAAGTTACCGGCTATATATTGATATATAGAACTAAGGGCTTGCGCAATCTTGGTGATCTGTTCCCAAACCTCACCGTGGTGCGAGGGATGCAGCTCTTCAAGGATTTCGCTCTTGTAATATTTGACAATGAACATCTAGAGGCGCTTGGACTGAATTCGTTGATGAAGATCGAGCGAGGAGGCGTCCGTATTCAGCACAACGATCGACTCTGCTATACTAACACCATAGACTGGTCGCGAATCACACGCGATGATGCTGAAATCATCGTCAGGTCTAACTATGATACCAGACTGTGCGGCTTGTGCCCCAACGCTCAGAGTCGCGTTGAGGAGGACCATCGGCAATCGTTGCCGCAATGTCCCGCTGACACCAAAGGGAAGCTGCTTTGCTGGGACGAGAAACACTGCCAAAAGATATGTCCGAGCGCTTGCGGCGACCACGGCTGTATGTCGAACGGCACGTGCTGCAACTCCGCCTGTCTCGGAGGATGCGACGGACCGCTCGCGAGCAATTGCTTCGTCTGCAAGAAGTTCTCGTTTGAATACGGGTCCGAGATGACTTGTATGGAGTCCTGCCCCGGTGGTACTTACGAGTTGTCCCGGAGATGTGTGACGGAACAGGAGTGTCGCTCGATGTCCCCCCCGCCGCAGCCGGTCGGCGCCGCGGACTCCACGTACTTCCGGTTCGCGATCAAGGTGCCCGCCTATAAGACATTCAACAGCAGCCGCTGTCTGTTCATGTGTCCGAGTGGCTACATGGAGGTGGGTAACGAGACTAACGCGACGTGCCAGCCGTGCCCGCGGTCCGGCTGCGTGCGCGAGTGCCTCGGCGGGAAGATCGACTCGGTCGCCACCGCGGAGCAGTTCCGGGGCTGCACGCACATTAAGGGCAAAATGGAATTCACGCTTCGCTCCAGCGGAGGTACGTTTATACTGGACCATAGCACTGTTTCGAATTCAAGACATGAGGGATGA
Protein
MDLRNNPDQIRRLEGCRVIEGQLSIVLMERATNQIFDNMSFPQLREVTGYILIYRTKGLRNLGDLFPNLTVVRGMQLFKDFALVIFDNEHLEALGLNSLMKIERGGVRIQHNDRLCYTNTIDWSRITRDDAEIIVRSNYDTRLCGLCPNAQSRVEEDHRQSLPQCPADTKGKLLCWDEKHCQKICPSACGDHGCMSNGTCCNSACLGGCDGPLASNCFVCKKFSFEYGSEMTCMESCPGGTYELSRRCVTEQECRSMSPPPQPVGAADSTYFRFAIKVPAYKTFNSSRCLFMCPSGYMEVGNETNATCQPCPRSGCVRECLGGKIDSVATAEQFRGCTHIKGKMEFTLRSSGGTFILDHSTVSNSRHEG
Summary
Description
This receptor probably binds an insulin related protein and has a tyrosine-protein kinase activity.
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Cofactor
Mn(2+)
Subunit
Tetramer of 2 alpha and 2 beta chains linked by disulfide bonds. The alpha chains contribute to the formation of the ligand-binding domain, while the beta chains carry the kinase domain (By similarity).
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family. Insulin receptor subfamily.
Keywords
ATP-binding
Cleavage on pair of basic residues
Complete proteome
Disulfide bond
Glycoprotein
Kinase
Manganese
Membrane
Metal-binding
Nucleotide-binding
Phosphoprotein
Receptor
Reference proteome
Repeat
Signal
Transferase
Transmembrane
Transmembrane helix
Tyrosine-protein kinase
Feature
chain Tyrosine-protein kinase receptor
Uniprot
H9J2B7
Q9U5A8
A0A2H1W0I4
A0A2A4JUH3
A0A2W1BQ19
A0A212ELK6
+ More
A0A194PNX0 A0A2D3E1C8 A0A194QUL6 A0A1B6FCK7 E2A530 A0A1B6KAQ3 A0A2J7R3R6 A0A195CNS3 A0A151JYW9 A0A195BAD2 A0A158NFV3 A0A151X3W9 A0A3L8D597 A0A151IYJ9 A0A067QUW8 A0A0C9QS61 A0A0G4DDB6 A0A1L8DJW7 G3ER33 A0A2S2R3F4 A0A1Y1KXL1 W4VRQ3 A0A088AUM7 G2J5R2 A0A2R4G8S9 A0A1Q3G5F2 A0A1Q3G5E9 A0A0C4UQS4 A0A2R4G8T0 A0A1B6DYS9 A0A1Q3G5E6 A0A1Q3G5E7 A0A1Q3G5I4 A0A1Q3G5E5 A0A2P8YHI3 A0A182JT77 A0A182MVW1 A0A1B1UZE9 A0A1W4XBR9 A0A2S2NHA9 A0A1B0C601 A0A146L9I6 A0A182Y1F3 A0A1B0AJ67 A0A146KY69 R4IJT3 A0A2S2N7Z7 A0A2M4A0Z5 A0A1A9UWD9 A0A2M4A0A1 A0A2M4A0F2 A0A2H8TMN4 A0A182RD25 A0A2M4A3E1 A0A310SXM6 A0A2M4A092 A0A182WEW1 A0A0L7R7M6 A0A0A9YBQ5 J9K8T8 A0A1A9YHT5 A0A1V0JFF9 Q7Q0F6 A0A0B4L064 D6W7P5 A0A0N0BDV4 A0A182FBB5 A0A1J1J845 Q93105 W8BST3 A0A2P1CYK5 A0A1S4F1J9 A0A034UZV1 A0A0K8V795 A0A0K8TY52 A0A0K0PQK7 W5JGL8 A0A0T6BFY0 A0A2M3YYA3 A0A182MYU2 A0A2M4B938 A0A2M4B8W8 A0A2R7WTX2 A0A1A9WJ29 N6T8W2 U4UVJ9 A0A182QRS4 A0A0A1WF79 K9M126 T1D5Z9 A0A0P5CRI0 V9KI02 A0A0P5EVH0 A0A336M6T9
A0A194PNX0 A0A2D3E1C8 A0A194QUL6 A0A1B6FCK7 E2A530 A0A1B6KAQ3 A0A2J7R3R6 A0A195CNS3 A0A151JYW9 A0A195BAD2 A0A158NFV3 A0A151X3W9 A0A3L8D597 A0A151IYJ9 A0A067QUW8 A0A0C9QS61 A0A0G4DDB6 A0A1L8DJW7 G3ER33 A0A2S2R3F4 A0A1Y1KXL1 W4VRQ3 A0A088AUM7 G2J5R2 A0A2R4G8S9 A0A1Q3G5F2 A0A1Q3G5E9 A0A0C4UQS4 A0A2R4G8T0 A0A1B6DYS9 A0A1Q3G5E6 A0A1Q3G5E7 A0A1Q3G5I4 A0A1Q3G5E5 A0A2P8YHI3 A0A182JT77 A0A182MVW1 A0A1B1UZE9 A0A1W4XBR9 A0A2S2NHA9 A0A1B0C601 A0A146L9I6 A0A182Y1F3 A0A1B0AJ67 A0A146KY69 R4IJT3 A0A2S2N7Z7 A0A2M4A0Z5 A0A1A9UWD9 A0A2M4A0A1 A0A2M4A0F2 A0A2H8TMN4 A0A182RD25 A0A2M4A3E1 A0A310SXM6 A0A2M4A092 A0A182WEW1 A0A0L7R7M6 A0A0A9YBQ5 J9K8T8 A0A1A9YHT5 A0A1V0JFF9 Q7Q0F6 A0A0B4L064 D6W7P5 A0A0N0BDV4 A0A182FBB5 A0A1J1J845 Q93105 W8BST3 A0A2P1CYK5 A0A1S4F1J9 A0A034UZV1 A0A0K8V795 A0A0K8TY52 A0A0K0PQK7 W5JGL8 A0A0T6BFY0 A0A2M3YYA3 A0A182MYU2 A0A2M4B938 A0A2M4B8W8 A0A2R7WTX2 A0A1A9WJ29 N6T8W2 U4UVJ9 A0A182QRS4 A0A0A1WF79 K9M126 T1D5Z9 A0A0P5CRI0 V9KI02 A0A0P5EVH0 A0A336M6T9
EC Number
2.7.10.1
Pubmed
19121390
28756777
22118469
26354079
20798317
21347285
+ More
30249741 24845553 24657890 21797944 28004739 25799997 29403074 26823975 25244985 23499946 25401762 28230772 12364791 18362917 19820115 9099579 17510324 24495485 25348373 26812356 20920257 23761445 23537049 25830018 23136112 24330624 24402279
30249741 24845553 24657890 21797944 28004739 25799997 29403074 26823975 25244985 23499946 25401762 28230772 12364791 18362917 19820115 9099579 17510324 24495485 25348373 26812356 20920257 23761445 23537049 25830018 23136112 24330624 24402279
EMBL
BABH01007614
BABH01007615
BABH01007616
AF025542
AAF21243.1
ODYU01005566
+ More
SOQ46537.1 NWSH01000609 PCG75348.1 KZ149960 PZC76371.1 AGBW02014037 OWR42372.1 KQ459597 KPI95007.1 MF443095 ATU31392.1 KQ461150 KPJ08670.1 GECZ01022086 JAS47683.1 GL436794 EFN71461.1 GEBQ01031456 JAT08521.1 NEVH01007818 PNF35477.1 KQ977481 KYN02366.1 KQ981463 KYN41515.1 KQ976532 KYM81501.1 ADTU01000013 ADTU01000014 ADTU01000015 KQ982557 KYQ55071.1 QOIP01000012 RLU15667.1 KQ980763 KYN13503.1 KK852921 KDR13786.1 GBYB01006589 JAG76356.1 HG518668 CDI30232.1 GFDF01007444 JAV06640.1 JF304722 ADZ56366.1 GGMS01015326 MBY84529.1 GEZM01075366 JAV64116.1 GANO01000511 JAB59360.1 BK008012 DAA34971.1 MF620042 AVT56264.1 GFDL01000018 JAV35027.1 GFDL01000017 JAV35028.1 KF974333 AIY24638.1 MF620044 AVT56266.1 GEDC01006489 JAS30809.1 GFDL01000014 JAV35031.1 GFDL01000013 JAV35032.1 GFDL01000016 JAV35029.1 GFDL01000015 JAV35030.1 PYGN01000591 PSN43728.1 AXCM01007680 KX428482 ANW09593.1 GGMR01003889 MBY16508.1 JXJN01026385 GDHC01013621 JAQ05008.1 GDHC01018543 GDHC01006354 JAQ00086.1 JAQ12275.1 JX291127 AFQ01096.1 GGMR01000678 MBY13297.1 GGFK01000987 MBW34308.1 GGFK01000923 MBW34244.1 GGFK01000963 MBW34284.1 GFXV01003618 MBW15423.1 GGFK01002015 MBW35336.1 KQ759862 OAD62524.1 GGFK01000819 MBW34140.1 KQ414638 KOC66875.1 GBHO01041436 GBHO01014558 GBRD01008020 JAG02168.1 JAG29046.1 JAG57801.1 ABLF02025445 ABLF02025449 KX507134 ARD07921.1 AAAB01008986 EAA00322.3 KF922648 AHF20214.1 KQ971307 EFA11583.2 KQ435850 KOX70962.1 CVRI01000074 CRL08130.1 U72939 CH477238 GAMC01014291 JAB92264.1 KY992933 AVK43099.1 GAKP01022416 JAC36536.1 GDHF01017583 GDHF01012251 GDHF01000334 JAI34731.1 JAI40063.1 JAI51980.1 GDHF01033113 JAI19201.1 KP331063 KT355584 AKQ48960.1 ALV82503.1 ADMH02001279 ETN63231.1 LJIG01000694 KRT86246.1 GGFM01000502 MBW21253.1 GGFJ01000379 MBW49520.1 GGFJ01000338 MBW49479.1 KK855531 PTY22998.1 APGK01047122 KB741077 ENN74143.1 KB632384 ERL94276.1 AXCN02001027 GBXI01017152 GBXI01014387 GBXI01010478 JAC97139.1 JAC99904.1 JAD03814.1 JN711464 AFQ20827.1 GALA01000303 JAA94549.1 GDIP01167714 JAJ55688.1 JW865004 AFO97521.1 GDIQ01271234 JAJ80490.1 UFQT01000641 SSX26014.1
SOQ46537.1 NWSH01000609 PCG75348.1 KZ149960 PZC76371.1 AGBW02014037 OWR42372.1 KQ459597 KPI95007.1 MF443095 ATU31392.1 KQ461150 KPJ08670.1 GECZ01022086 JAS47683.1 GL436794 EFN71461.1 GEBQ01031456 JAT08521.1 NEVH01007818 PNF35477.1 KQ977481 KYN02366.1 KQ981463 KYN41515.1 KQ976532 KYM81501.1 ADTU01000013 ADTU01000014 ADTU01000015 KQ982557 KYQ55071.1 QOIP01000012 RLU15667.1 KQ980763 KYN13503.1 KK852921 KDR13786.1 GBYB01006589 JAG76356.1 HG518668 CDI30232.1 GFDF01007444 JAV06640.1 JF304722 ADZ56366.1 GGMS01015326 MBY84529.1 GEZM01075366 JAV64116.1 GANO01000511 JAB59360.1 BK008012 DAA34971.1 MF620042 AVT56264.1 GFDL01000018 JAV35027.1 GFDL01000017 JAV35028.1 KF974333 AIY24638.1 MF620044 AVT56266.1 GEDC01006489 JAS30809.1 GFDL01000014 JAV35031.1 GFDL01000013 JAV35032.1 GFDL01000016 JAV35029.1 GFDL01000015 JAV35030.1 PYGN01000591 PSN43728.1 AXCM01007680 KX428482 ANW09593.1 GGMR01003889 MBY16508.1 JXJN01026385 GDHC01013621 JAQ05008.1 GDHC01018543 GDHC01006354 JAQ00086.1 JAQ12275.1 JX291127 AFQ01096.1 GGMR01000678 MBY13297.1 GGFK01000987 MBW34308.1 GGFK01000923 MBW34244.1 GGFK01000963 MBW34284.1 GFXV01003618 MBW15423.1 GGFK01002015 MBW35336.1 KQ759862 OAD62524.1 GGFK01000819 MBW34140.1 KQ414638 KOC66875.1 GBHO01041436 GBHO01014558 GBRD01008020 JAG02168.1 JAG29046.1 JAG57801.1 ABLF02025445 ABLF02025449 KX507134 ARD07921.1 AAAB01008986 EAA00322.3 KF922648 AHF20214.1 KQ971307 EFA11583.2 KQ435850 KOX70962.1 CVRI01000074 CRL08130.1 U72939 CH477238 GAMC01014291 JAB92264.1 KY992933 AVK43099.1 GAKP01022416 JAC36536.1 GDHF01017583 GDHF01012251 GDHF01000334 JAI34731.1 JAI40063.1 JAI51980.1 GDHF01033113 JAI19201.1 KP331063 KT355584 AKQ48960.1 ALV82503.1 ADMH02001279 ETN63231.1 LJIG01000694 KRT86246.1 GGFM01000502 MBW21253.1 GGFJ01000379 MBW49520.1 GGFJ01000338 MBW49479.1 KK855531 PTY22998.1 APGK01047122 KB741077 ENN74143.1 KB632384 ERL94276.1 AXCN02001027 GBXI01017152 GBXI01014387 GBXI01010478 JAC97139.1 JAC99904.1 JAD03814.1 JN711464 AFQ20827.1 GALA01000303 JAA94549.1 GDIP01167714 JAJ55688.1 JW865004 AFO97521.1 GDIQ01271234 JAJ80490.1 UFQT01000641 SSX26014.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000000311
+ More
UP000235965 UP000078542 UP000078541 UP000078540 UP000005205 UP000075809 UP000279307 UP000078492 UP000027135 UP000005203 UP000245037 UP000075881 UP000075883 UP000192223 UP000092460 UP000076408 UP000092445 UP000078200 UP000075900 UP000075920 UP000053825 UP000007819 UP000092443 UP000007062 UP000007266 UP000053105 UP000069272 UP000183832 UP000008820 UP000000673 UP000075884 UP000091820 UP000019118 UP000030742 UP000075886
UP000235965 UP000078542 UP000078541 UP000078540 UP000005205 UP000075809 UP000279307 UP000078492 UP000027135 UP000005203 UP000245037 UP000075881 UP000075883 UP000192223 UP000092460 UP000076408 UP000092445 UP000078200 UP000075900 UP000075920 UP000053825 UP000007819 UP000092443 UP000007062 UP000007266 UP000053105 UP000069272 UP000183832 UP000008820 UP000000673 UP000075884 UP000091820 UP000019118 UP000030742 UP000075886
Interpro
IPR002011
Tyr_kinase_rcpt_2_CS
+ More
IPR006212 Furin_repeat
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR000494 Rcpt_L-dom
IPR006211 Furin-like_Cys-rich_dom
IPR036941 Rcpt_L-dom_sf
IPR000719 Prot_kinase_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR036116 FN3_sf
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR013783 Ig-like_fold
IPR016246 Tyr_kinase_insulin-like_rcpt
IPR009030 Growth_fac_rcpt_cys_sf
IPR003961 FN3_dom
IPR036034 PDZ_sf
IPR001478 PDZ
IPR028792 INSRR
IPR017896 4Fe4S_Fe-S-bd
IPR006212 Furin_repeat
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR000494 Rcpt_L-dom
IPR006211 Furin-like_Cys-rich_dom
IPR036941 Rcpt_L-dom_sf
IPR000719 Prot_kinase_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR036116 FN3_sf
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR013783 Ig-like_fold
IPR016246 Tyr_kinase_insulin-like_rcpt
IPR009030 Growth_fac_rcpt_cys_sf
IPR003961 FN3_dom
IPR036034 PDZ_sf
IPR001478 PDZ
IPR028792 INSRR
IPR017896 4Fe4S_Fe-S-bd
Gene 3D
ProteinModelPortal
H9J2B7
Q9U5A8
A0A2H1W0I4
A0A2A4JUH3
A0A2W1BQ19
A0A212ELK6
+ More
A0A194PNX0 A0A2D3E1C8 A0A194QUL6 A0A1B6FCK7 E2A530 A0A1B6KAQ3 A0A2J7R3R6 A0A195CNS3 A0A151JYW9 A0A195BAD2 A0A158NFV3 A0A151X3W9 A0A3L8D597 A0A151IYJ9 A0A067QUW8 A0A0C9QS61 A0A0G4DDB6 A0A1L8DJW7 G3ER33 A0A2S2R3F4 A0A1Y1KXL1 W4VRQ3 A0A088AUM7 G2J5R2 A0A2R4G8S9 A0A1Q3G5F2 A0A1Q3G5E9 A0A0C4UQS4 A0A2R4G8T0 A0A1B6DYS9 A0A1Q3G5E6 A0A1Q3G5E7 A0A1Q3G5I4 A0A1Q3G5E5 A0A2P8YHI3 A0A182JT77 A0A182MVW1 A0A1B1UZE9 A0A1W4XBR9 A0A2S2NHA9 A0A1B0C601 A0A146L9I6 A0A182Y1F3 A0A1B0AJ67 A0A146KY69 R4IJT3 A0A2S2N7Z7 A0A2M4A0Z5 A0A1A9UWD9 A0A2M4A0A1 A0A2M4A0F2 A0A2H8TMN4 A0A182RD25 A0A2M4A3E1 A0A310SXM6 A0A2M4A092 A0A182WEW1 A0A0L7R7M6 A0A0A9YBQ5 J9K8T8 A0A1A9YHT5 A0A1V0JFF9 Q7Q0F6 A0A0B4L064 D6W7P5 A0A0N0BDV4 A0A182FBB5 A0A1J1J845 Q93105 W8BST3 A0A2P1CYK5 A0A1S4F1J9 A0A034UZV1 A0A0K8V795 A0A0K8TY52 A0A0K0PQK7 W5JGL8 A0A0T6BFY0 A0A2M3YYA3 A0A182MYU2 A0A2M4B938 A0A2M4B8W8 A0A2R7WTX2 A0A1A9WJ29 N6T8W2 U4UVJ9 A0A182QRS4 A0A0A1WF79 K9M126 T1D5Z9 A0A0P5CRI0 V9KI02 A0A0P5EVH0 A0A336M6T9
A0A194PNX0 A0A2D3E1C8 A0A194QUL6 A0A1B6FCK7 E2A530 A0A1B6KAQ3 A0A2J7R3R6 A0A195CNS3 A0A151JYW9 A0A195BAD2 A0A158NFV3 A0A151X3W9 A0A3L8D597 A0A151IYJ9 A0A067QUW8 A0A0C9QS61 A0A0G4DDB6 A0A1L8DJW7 G3ER33 A0A2S2R3F4 A0A1Y1KXL1 W4VRQ3 A0A088AUM7 G2J5R2 A0A2R4G8S9 A0A1Q3G5F2 A0A1Q3G5E9 A0A0C4UQS4 A0A2R4G8T0 A0A1B6DYS9 A0A1Q3G5E6 A0A1Q3G5E7 A0A1Q3G5I4 A0A1Q3G5E5 A0A2P8YHI3 A0A182JT77 A0A182MVW1 A0A1B1UZE9 A0A1W4XBR9 A0A2S2NHA9 A0A1B0C601 A0A146L9I6 A0A182Y1F3 A0A1B0AJ67 A0A146KY69 R4IJT3 A0A2S2N7Z7 A0A2M4A0Z5 A0A1A9UWD9 A0A2M4A0A1 A0A2M4A0F2 A0A2H8TMN4 A0A182RD25 A0A2M4A3E1 A0A310SXM6 A0A2M4A092 A0A182WEW1 A0A0L7R7M6 A0A0A9YBQ5 J9K8T8 A0A1A9YHT5 A0A1V0JFF9 Q7Q0F6 A0A0B4L064 D6W7P5 A0A0N0BDV4 A0A182FBB5 A0A1J1J845 Q93105 W8BST3 A0A2P1CYK5 A0A1S4F1J9 A0A034UZV1 A0A0K8V795 A0A0K8TY52 A0A0K0PQK7 W5JGL8 A0A0T6BFY0 A0A2M3YYA3 A0A182MYU2 A0A2M4B938 A0A2M4B8W8 A0A2R7WTX2 A0A1A9WJ29 N6T8W2 U4UVJ9 A0A182QRS4 A0A0A1WF79 K9M126 T1D5Z9 A0A0P5CRI0 V9KI02 A0A0P5EVH0 A0A336M6T9
PDB
6HN5
E-value=2.82804e-62,
Score=605
Ontologies
PATHWAY
GO
PANTHER
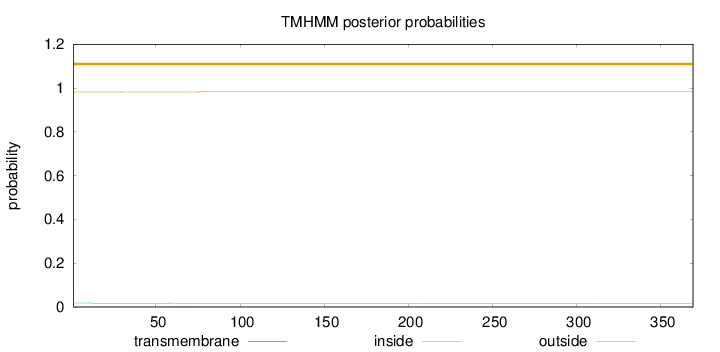
Topology
Subcellular location
Membrane
Length:
369
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01266
Exp number, first 60 AAs:
0.00453
Total prob of N-in:
0.01774
outside
1 - 369
Population Genetic Test Statistics
Pi
198.843423
Theta
175.166996
Tajima's D
0.547384
CLR
0.887191
CSRT
0.528123593820309
Interpretation
Uncertain
