Gene
KWMTBOMO02890
Pre Gene Modal
BGIBMGA003654
Annotation
PREDICTED:_insulin_receptor_isoform_X1_[Bombyx_mori]
Full name
Tyrosine-protein kinase receptor
Location in the cell
Cytoplasmic Reliability : 1.308
Sequence
CDS
ATGTTTTTGAAGAAACTCAGGAAAATTATCGCGAATCCTTCAGAAGATAAAGGGGAAGCTCTGCATATAATAAACAACGAGAATCTAGAGCTGCTATGGGACTGGTCCACTTACGGACCTATAGAGATAATCGGTAGCTGCTACATACATTCGAATCCCAAGCTGTGCTACACTCAACTGATGCCGCTTATGAATATGTCGTATCCGAAAACGAATTTTACCGAACTAGAAGTGTCCCAAGACAGCAACGGGTATCAGGCATCTTGTCTCCCGGACGTCCTGAAGCTGAGCGTGTCGGAGTTAACGCAAATCGCAGTGGTACTAACTTGGGATGTGTACTGTCCCGACGACATGCGCAAGCTGCTCGGATACTCGCTGTACTTTATGGCCACCGAGAAGAACGTAACTCTGTACGATCAACGCGATGCCTGCTCTGATATATGGAAAGTACTCGACAAGCCAATAGAGGAGACTCGTAAAGTGAACCGCAACGCGAAGCCCTCTCACGCCAGGAACAAAACGGAGGTTATATTCAATCCGTGCGCCGACATGCAGCCGCTGTTCTATCCCATCACCCAGCTGAGGCCATACACCAGATACGCGGCCTACGTGAAGACCTACACCACGATACAGGACAGGAAGGGGGCCCAGAGCTCGATCATTTACTTCAGGACCTTGCCGATGCAACCGAGTCCACCCACCGCGGTCACTGTGGAGCTGTTGACGCCGCATTCGATCTTCATAAAATGGTCACCGCCGACGATGCCGAACGGTACGATCATACTGTACCACGTGGAAGTGCAAGCCAACAGCTACAACAGGCCGCAGATACTTGCCGGCAACATCAATTATTGTGGCAATCCGAGCATCTTGGCTAACATGATCAAAGCTAGTTTAGAAGAAGCGCCCGAAACTCCAGAGGAGAAAACCACCGGCGACGTGAAAAATGGTTTTTGCGCTTGCAAAGAGGAAGCCAAACCGAGTCTGAGGTTCAGCACTCGCGCCGAGGAGGAGAGGGTCGACAGCATCAATTTCGAGAACGAGTTGCAAAATCAGGTTTACAGGAAATCCGAACGAGTGAACCCGAAAATACACAAGTCTATCGACGGAACGAGCAAAGTGAAAAGGTCGGTCGACCACAGCCTCACCAGCATGCTCGTCAAACTGACCGAGAACAACAGCCCGAGACCCGGCCTCAACTACTCCAACATCACGGACGACGAAGGTTACGTGAAATCTTTATACTACGAATTGGACGGCTCGACTCGGACGTTGACCGTGAAGAACATGCGTCACTTCACCTGGTACACCGTGAACATATGGGCGTGTCGCAACAAGGACTTGAACGAAACCGATCCGGTCTACGCCGAGAACTGGTGCTCGGTTCGCGCTTATAACACGTTCCGCACCTTAGAACTGCCAAACGCAGACGTTGTGAGCAACTTCCAAGTGGAGATAATGCCGGCTAACAAGACCTTGCCCGAAGTGAACGTCACGTGGGAGCCTCCCCTCAATCCGAACGGTTTTGTGGTCGCGTACAACGTACATTACTCACGAATCGAGAACAGTGCGCAGGGCCCGGATGTAGGTTTACAGAGCTGTATAACAGTAGACGACTACGAAGCGAACGGGCACGGCTACGTGCTCAAGACCCTATCTCCGGGCAACTACAGCGTCAAGGTCACACCGGTCACCGTCAGCGGAGCAGGAAACGTTTCCGAAGAAATCTACGTATTTATACCGGAGCGAGTATCGGAGAGCGGGCACGCGTGGGCGTGGGGCGTGGGGGCGGGCGGCGTGGTGCTGGCGCTGCTGGTGGGCGCGGCCGTGTGGTACGCGCGCCGCGGCCTCATGTCCCACGCCGAGGCGAGCAAGCTCTTCGCCAGCGTCAACCCGGAGTACGTGCCCACTGTCTACGTGCCCGACGAGTGGGAGGTCACACGCGACAGCATACACTTCATCCGAGAACTCGGACAGGGATCGTTTGGCATGGTGTATGAAGGGATCGCCAAGGAAATCGTGAAAGGGAAACCGGAAACGCGCTGCGCCATCAAAACCGTTAACGAGCACGCGACAGACAGGGAGCGCATCGAGTTCCTGAACGAGGCGTCGGTGATGAAGGCGTTTGACACGTTCCACGTGGTGCGGCTGCTGGGCGTGGTCTCCCGCGGGCAGCCGACGCTCGTCGTGATGGAGCTCATGGAGCGCGGCGACCTCAAGACGTACCTGCGCTCCATGCGACCCGACGCCGAGGGCTCGCTGCCCTCCAGCCCGCCCGTGCCGCCCACGCTGCAGAACATCTTGCAGATGGCCATAGAAATAGCCGATGGGATGGCATATTTGTCGGCAAAAAAATTCGTGCACCGCGACTTAGCGGCGCGCAACTGTATGGTCGCCGGCGACCTCACCGTCAAAGTTGGGGACTTCGGCATGACCAGAGATATTTATGAGACTGATTATTATAGGAAAGGCACCAAAGGCCTAATGCCGGTGCGGTGGATGAGCCCCGAAAGCTTAAAAGACGGGGTCTTTTCCAGTAGCAGTGACGCTTGGAGTTACGGGGTTGTTTTGTGGGAGATGGCAACGCTCGCGATGCAACCCTATCAGGGGTTGTCGAACGAGCAGGTGCTGCGGTACGTGGTGGAGGGCGGGGTCATGGAGCGGCCGGAGCAGTGCCCGGACCGGCTGTACGAGCTCATGCGCGCGTGCTGGACGCACCGCCCCTCCGCGCGCCCCACCTTCCTGCAGCTGGTGGCCGACCTGGTGCCGCACGCCATGCCCTACTTCCGCCACCGCAGCTTCTTCCACTCGCCGCAGGGCCAGGAGTTGTACGCTCTCCAGCGATCTGCGCTAGATGAGGAACAAGAGCTCCCGGAGGTGGACGTGGGTGCGGTGGCGACGGGCTCCGTGTCGAACCTGTTCGGTGTGTCGGGGCGGCTCGCGTCCTGGGTGCGGGAGCTGTCCTCTCTGCGGTCCCGGGGCTCCGACGACGCGGCGGCCGAGCCTCTGCAGCCCCTCCCTCCGCCCGGCACCTCCCCCGCCCCCATACTCGCACCCGCACCGGCCATCAAGGGACCCAACGGCGTACTGCGCGACTCGCACTCTGCCCGTTTTCGAAACCACTGA
Protein
MFLKKLRKIIANPSEDKGEALHIINNENLELLWDWSTYGPIEIIGSCYIHSNPKLCYTQLMPLMNMSYPKTNFTELEVSQDSNGYQASCLPDVLKLSVSELTQIAVVLTWDVYCPDDMRKLLGYSLYFMATEKNVTLYDQRDACSDIWKVLDKPIEETRKVNRNAKPSHARNKTEVIFNPCADMQPLFYPITQLRPYTRYAAYVKTYTTIQDRKGAQSSIIYFRTLPMQPSPPTAVTVELLTPHSIFIKWSPPTMPNGTIILYHVEVQANSYNRPQILAGNINYCGNPSILANMIKASLEEAPETPEEKTTGDVKNGFCACKEEAKPSLRFSTRAEEERVDSINFENELQNQVYRKSERVNPKIHKSIDGTSKVKRSVDHSLTSMLVKLTENNSPRPGLNYSNITDDEGYVKSLYYELDGSTRTLTVKNMRHFTWYTVNIWACRNKDLNETDPVYAENWCSVRAYNTFRTLELPNADVVSNFQVEIMPANKTLPEVNVTWEPPLNPNGFVVAYNVHYSRIENSAQGPDVGLQSCITVDDYEANGHGYVLKTLSPGNYSVKVTPVTVSGAGNVSEEIYVFIPERVSESGHAWAWGVGAGGVVLALLVGAAVWYARRGLMSHAEASKLFASVNPEYVPTVYVPDEWEVTRDSIHFIRELGQGSFGMVYEGIAKEIVKGKPETRCAIKTVNEHATDRERIEFLNEASVMKAFDTFHVVRLLGVVSRGQPTLVVMELMERGDLKTYLRSMRPDAEGSLPSSPPVPPTLQNILQMAIEIADGMAYLSAKKFVHRDLAARNCMVAGDLTVKVGDFGMTRDIYETDYYRKGTKGLMPVRWMSPESLKDGVFSSSSDAWSYGVVLWEMATLAMQPYQGLSNEQVLRYVVEGGVMERPEQCPDRLYELMRACWTHRPSARPTFLQLVADLVPHAMPYFRHRSFFHSPQGQELYALQRSALDEEQELPEVDVGAVATGSVSNLFGVSGRLASWVRELSSLRSRGSDDAAAEPLQPLPPPGTSPAPILAPAPAIKGPNGVLRDSHSARFRNH
Summary
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family. Insulin receptor subfamily.
Feature
chain Tyrosine-protein kinase receptor
Uniprot
Q9U5A8
H9J2B7
B5U451
A0A2A4JUH3
A0A2W1BQ19
A0A194QYL1
+ More
A0A194PNX0 A0A212ELK6 A0A2D3E1C8 A0A2J7R3R8 A0A2J7R3R6 A0A1Y1KXL1 A0A1W4XBR9 A0A1B1UZE9 G3ER33 F4WE54 E2A530 A0A151X3W9 A0A151IYJ9 A0A0J7KR93 A0A195BAD2 A0A158NFV3 A0A067QUW8 A0A195CNS3 A0A2P1CYK5 A0A1B6KAQ3 E2B4Y3 A0A310SXM6 A0A0K0PQK7 A0A0C9QS61 A0A3L8D597 G2J5R2 A0A088AUM7 A0A026W1L5 A0A0N0BDV4 A0A2A3ESY9 A0A2R4G8S9 A0A2R4G8T0 A0A0L7R7M6 A0A2D0RTI5 A0A1A9WJ29 K7F279 K7F278 A0A1A9YHT5 W5M9H7 A0A3B3SIB8 W5M9K1 A0A087Y256 A0A3B3Z515 A0A2M4B8W8 A0A0K0QRF3 M4AW90 A0A2M4B938 A0A146P5T1 A0A146P740 A0A075IJV4 A0A096M0W0 I3KQM7 A0A1A7Y0I0 A0A3B5PVH2 A0A096M5E1 A0A087XET4 A0A3P9DDV5 A0A1A8C426 A0A3B1J7U9 A0A3B3YP57 A0A1A8JWQ8 A0A3B3YNW5 A0A3B4FJD9 F1QNX5 B7SNG4 A0A1A8QE15 A0A2M3YYU6 A0A1A8L188 A0A1S3R4V3 A0A2M3YYA3 A0A3P8PHL2 G3WLB9 A0A146N073 A0A3B3VRB6 A0A1A7ZXN0 G1T855 G1TLR2 F1PXU6 A0A076FVY1 I3MHA3 A0A384BUF8 A0A2I4AKQ2 A0A3P8W3R3 G1PWB2 U6D754 A0A250Y7G3 G1LGG8 A0A2Y9GBI4 M3WPQ6 A0A2U3ZLZ8 A0A2U9C2U4 A0A091KYJ0
A0A194PNX0 A0A212ELK6 A0A2D3E1C8 A0A2J7R3R8 A0A2J7R3R6 A0A1Y1KXL1 A0A1W4XBR9 A0A1B1UZE9 G3ER33 F4WE54 E2A530 A0A151X3W9 A0A151IYJ9 A0A0J7KR93 A0A195BAD2 A0A158NFV3 A0A067QUW8 A0A195CNS3 A0A2P1CYK5 A0A1B6KAQ3 E2B4Y3 A0A310SXM6 A0A0K0PQK7 A0A0C9QS61 A0A3L8D597 G2J5R2 A0A088AUM7 A0A026W1L5 A0A0N0BDV4 A0A2A3ESY9 A0A2R4G8S9 A0A2R4G8T0 A0A0L7R7M6 A0A2D0RTI5 A0A1A9WJ29 K7F279 K7F278 A0A1A9YHT5 W5M9H7 A0A3B3SIB8 W5M9K1 A0A087Y256 A0A3B3Z515 A0A2M4B8W8 A0A0K0QRF3 M4AW90 A0A2M4B938 A0A146P5T1 A0A146P740 A0A075IJV4 A0A096M0W0 I3KQM7 A0A1A7Y0I0 A0A3B5PVH2 A0A096M5E1 A0A087XET4 A0A3P9DDV5 A0A1A8C426 A0A3B1J7U9 A0A3B3YP57 A0A1A8JWQ8 A0A3B3YNW5 A0A3B4FJD9 F1QNX5 B7SNG4 A0A1A8QE15 A0A2M3YYU6 A0A1A8L188 A0A1S3R4V3 A0A2M3YYA3 A0A3P8PHL2 G3WLB9 A0A146N073 A0A3B3VRB6 A0A1A7ZXN0 G1T855 G1TLR2 F1PXU6 A0A076FVY1 I3MHA3 A0A384BUF8 A0A2I4AKQ2 A0A3P8W3R3 G1PWB2 U6D754 A0A250Y7G3 G1LGG8 A0A2Y9GBI4 M3WPQ6 A0A2U3ZLZ8 A0A2U9C2U4 A0A091KYJ0
EC Number
2.7.10.1
Pubmed
EMBL
AF025542
AAF21243.1
BABH01007614
BABH01007615
BABH01007616
FJ169464
+ More
ACI02334.1 NWSH01000609 PCG75348.1 KZ149960 PZC76371.1 KQ461150 KPJ08671.1 KQ459597 KPI95007.1 AGBW02014037 OWR42372.1 MF443095 ATU31392.1 NEVH01007818 PNF35478.1 PNF35477.1 GEZM01075366 JAV64116.1 KX428482 ANW09593.1 JF304722 ADZ56366.1 GL888102 EGI67492.1 GL436794 EFN71461.1 KQ982557 KYQ55071.1 KQ980763 KYN13503.1 LBMM01004093 KMQ92816.1 KQ976532 KYM81501.1 ADTU01000013 ADTU01000014 ADTU01000015 KK852921 KDR13786.1 KQ977481 KYN02366.1 KY992933 AVK43099.1 GEBQ01031456 JAT08521.1 GL445681 EFN89247.1 KQ759862 OAD62524.1 KP331063 KT355584 AKQ48960.1 ALV82503.1 GBYB01006589 JAG76356.1 QOIP01000012 RLU15667.1 BK008012 DAA34971.1 KK107558 EZA48949.1 KQ435850 KOX70962.1 KZ288192 PBC34259.1 MF620042 AVT56264.1 MF620044 AVT56266.1 KQ414638 KOC66875.1 AGCU01111388 AGCU01111389 AGCU01111390 AGCU01111391 AGCU01111392 AGCU01111393 AHAT01011134 AHAT01011135 AYCK01005051 AYCK01005052 AYCK01005053 GGFJ01000338 MBW49479.1 KP713801 AKR15643.1 GGFJ01000379 MBW49520.1 GCES01147150 JAQ39172.1 GCES01146635 JAQ39687.1 KF872769 AIF28000.1 AERX01019757 AERX01019758 HADW01016716 HADX01001268 SBP23500.1 AYCK01009878 AYCK01009879 AYCK01009880 HADZ01009499 SBP73440.1 HAED01020096 HAEE01004347 SBR24367.1 BX539340 EU447177 ACC77574.1 HAEH01003705 HAEI01005166 SBR92065.1 GGFM01000675 MBW21426.1 HAEF01001193 SBR38575.1 GGFM01000502 MBW21253.1 AEFK01007044 AEFK01007045 AEFK01007046 AEFK01007047 GCES01161962 JAQ24360.1 HADY01008541 SBP47026.1 AAEX03002256 AAEX03002257 AAEX03002258 AAEX03002259 KM190088 AII23383.1 AGTP01015962 AGTP01015963 AGTP01015964 AGTP01015965 AAPE02054813 AAPE02054814 AAPE02054815 HAAF01006597 CCP78421.1 GFFV01000373 JAV39572.1 ACTA01003557 ACTA01179548 ACTA01187546 ACTA01195544 AANG04000441 CP026254 AWP10941.1 KK763525 KFP45649.1
ACI02334.1 NWSH01000609 PCG75348.1 KZ149960 PZC76371.1 KQ461150 KPJ08671.1 KQ459597 KPI95007.1 AGBW02014037 OWR42372.1 MF443095 ATU31392.1 NEVH01007818 PNF35478.1 PNF35477.1 GEZM01075366 JAV64116.1 KX428482 ANW09593.1 JF304722 ADZ56366.1 GL888102 EGI67492.1 GL436794 EFN71461.1 KQ982557 KYQ55071.1 KQ980763 KYN13503.1 LBMM01004093 KMQ92816.1 KQ976532 KYM81501.1 ADTU01000013 ADTU01000014 ADTU01000015 KK852921 KDR13786.1 KQ977481 KYN02366.1 KY992933 AVK43099.1 GEBQ01031456 JAT08521.1 GL445681 EFN89247.1 KQ759862 OAD62524.1 KP331063 KT355584 AKQ48960.1 ALV82503.1 GBYB01006589 JAG76356.1 QOIP01000012 RLU15667.1 BK008012 DAA34971.1 KK107558 EZA48949.1 KQ435850 KOX70962.1 KZ288192 PBC34259.1 MF620042 AVT56264.1 MF620044 AVT56266.1 KQ414638 KOC66875.1 AGCU01111388 AGCU01111389 AGCU01111390 AGCU01111391 AGCU01111392 AGCU01111393 AHAT01011134 AHAT01011135 AYCK01005051 AYCK01005052 AYCK01005053 GGFJ01000338 MBW49479.1 KP713801 AKR15643.1 GGFJ01000379 MBW49520.1 GCES01147150 JAQ39172.1 GCES01146635 JAQ39687.1 KF872769 AIF28000.1 AERX01019757 AERX01019758 HADW01016716 HADX01001268 SBP23500.1 AYCK01009878 AYCK01009879 AYCK01009880 HADZ01009499 SBP73440.1 HAED01020096 HAEE01004347 SBR24367.1 BX539340 EU447177 ACC77574.1 HAEH01003705 HAEI01005166 SBR92065.1 GGFM01000675 MBW21426.1 HAEF01001193 SBR38575.1 GGFM01000502 MBW21253.1 AEFK01007044 AEFK01007045 AEFK01007046 AEFK01007047 GCES01161962 JAQ24360.1 HADY01008541 SBP47026.1 AAEX03002256 AAEX03002257 AAEX03002258 AAEX03002259 KM190088 AII23383.1 AGTP01015962 AGTP01015963 AGTP01015964 AGTP01015965 AAPE02054813 AAPE02054814 AAPE02054815 HAAF01006597 CCP78421.1 GFFV01000373 JAV39572.1 ACTA01003557 ACTA01179548 ACTA01187546 ACTA01195544 AANG04000441 CP026254 AWP10941.1 KK763525 KFP45649.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000235965
+ More
UP000192223 UP000007755 UP000000311 UP000075809 UP000078492 UP000036403 UP000078540 UP000005205 UP000027135 UP000078542 UP000008237 UP000279307 UP000005203 UP000053097 UP000053105 UP000242457 UP000053825 UP000221080 UP000091820 UP000007267 UP000092443 UP000018468 UP000261540 UP000028760 UP000261480 UP000002852 UP000005207 UP000265160 UP000018467 UP000261460 UP000000437 UP000087266 UP000265100 UP000007648 UP000261500 UP000001811 UP000002254 UP000005215 UP000261680 UP000192220 UP000265120 UP000001074 UP000008912 UP000248481 UP000011712 UP000245340 UP000246464
UP000192223 UP000007755 UP000000311 UP000075809 UP000078492 UP000036403 UP000078540 UP000005205 UP000027135 UP000078542 UP000008237 UP000279307 UP000005203 UP000053097 UP000053105 UP000242457 UP000053825 UP000221080 UP000091820 UP000007267 UP000092443 UP000018468 UP000261540 UP000028760 UP000261480 UP000002852 UP000005207 UP000265160 UP000018467 UP000261460 UP000000437 UP000087266 UP000265100 UP000007648 UP000261500 UP000001811 UP000002254 UP000005215 UP000261680 UP000192220 UP000265120 UP000001074 UP000008912 UP000248481 UP000011712 UP000245340 UP000246464
Pfam
Interpro
IPR001245
Ser-Thr/Tyr_kinase_cat_dom
+ More
IPR036116 FN3_sf
IPR017441 Protein_kinase_ATP_BS
IPR016246 Tyr_kinase_insulin-like_rcpt
IPR003961 FN3_dom
IPR006212 Furin_repeat
IPR008266 Tyr_kinase_AS
IPR000494 Rcpt_L-dom
IPR036941 Rcpt_L-dom_sf
IPR011009 Kinase-like_dom_sf
IPR013783 Ig-like_fold
IPR009030 Growth_fac_rcpt_cys_sf
IPR002011 Tyr_kinase_rcpt_2_CS
IPR020635 Tyr_kinase_cat_dom
IPR006211 Furin-like_Cys-rich_dom
IPR000719 Prot_kinase_dom
IPR017896 4Fe4S_Fe-S-bd
IPR036034 PDZ_sf
IPR001478 PDZ
IPR028792 INSRR
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR040969 Insulin_TMD
IPR036116 FN3_sf
IPR017441 Protein_kinase_ATP_BS
IPR016246 Tyr_kinase_insulin-like_rcpt
IPR003961 FN3_dom
IPR006212 Furin_repeat
IPR008266 Tyr_kinase_AS
IPR000494 Rcpt_L-dom
IPR036941 Rcpt_L-dom_sf
IPR011009 Kinase-like_dom_sf
IPR013783 Ig-like_fold
IPR009030 Growth_fac_rcpt_cys_sf
IPR002011 Tyr_kinase_rcpt_2_CS
IPR020635 Tyr_kinase_cat_dom
IPR006211 Furin-like_Cys-rich_dom
IPR000719 Prot_kinase_dom
IPR017896 4Fe4S_Fe-S-bd
IPR036034 PDZ_sf
IPR001478 PDZ
IPR028792 INSRR
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR040969 Insulin_TMD
SUPFAM
Gene 3D
ProteinModelPortal
Q9U5A8
H9J2B7
B5U451
A0A2A4JUH3
A0A2W1BQ19
A0A194QYL1
+ More
A0A194PNX0 A0A212ELK6 A0A2D3E1C8 A0A2J7R3R8 A0A2J7R3R6 A0A1Y1KXL1 A0A1W4XBR9 A0A1B1UZE9 G3ER33 F4WE54 E2A530 A0A151X3W9 A0A151IYJ9 A0A0J7KR93 A0A195BAD2 A0A158NFV3 A0A067QUW8 A0A195CNS3 A0A2P1CYK5 A0A1B6KAQ3 E2B4Y3 A0A310SXM6 A0A0K0PQK7 A0A0C9QS61 A0A3L8D597 G2J5R2 A0A088AUM7 A0A026W1L5 A0A0N0BDV4 A0A2A3ESY9 A0A2R4G8S9 A0A2R4G8T0 A0A0L7R7M6 A0A2D0RTI5 A0A1A9WJ29 K7F279 K7F278 A0A1A9YHT5 W5M9H7 A0A3B3SIB8 W5M9K1 A0A087Y256 A0A3B3Z515 A0A2M4B8W8 A0A0K0QRF3 M4AW90 A0A2M4B938 A0A146P5T1 A0A146P740 A0A075IJV4 A0A096M0W0 I3KQM7 A0A1A7Y0I0 A0A3B5PVH2 A0A096M5E1 A0A087XET4 A0A3P9DDV5 A0A1A8C426 A0A3B1J7U9 A0A3B3YP57 A0A1A8JWQ8 A0A3B3YNW5 A0A3B4FJD9 F1QNX5 B7SNG4 A0A1A8QE15 A0A2M3YYU6 A0A1A8L188 A0A1S3R4V3 A0A2M3YYA3 A0A3P8PHL2 G3WLB9 A0A146N073 A0A3B3VRB6 A0A1A7ZXN0 G1T855 G1TLR2 F1PXU6 A0A076FVY1 I3MHA3 A0A384BUF8 A0A2I4AKQ2 A0A3P8W3R3 G1PWB2 U6D754 A0A250Y7G3 G1LGG8 A0A2Y9GBI4 M3WPQ6 A0A2U3ZLZ8 A0A2U9C2U4 A0A091KYJ0
A0A194PNX0 A0A212ELK6 A0A2D3E1C8 A0A2J7R3R8 A0A2J7R3R6 A0A1Y1KXL1 A0A1W4XBR9 A0A1B1UZE9 G3ER33 F4WE54 E2A530 A0A151X3W9 A0A151IYJ9 A0A0J7KR93 A0A195BAD2 A0A158NFV3 A0A067QUW8 A0A195CNS3 A0A2P1CYK5 A0A1B6KAQ3 E2B4Y3 A0A310SXM6 A0A0K0PQK7 A0A0C9QS61 A0A3L8D597 G2J5R2 A0A088AUM7 A0A026W1L5 A0A0N0BDV4 A0A2A3ESY9 A0A2R4G8S9 A0A2R4G8T0 A0A0L7R7M6 A0A2D0RTI5 A0A1A9WJ29 K7F279 K7F278 A0A1A9YHT5 W5M9H7 A0A3B3SIB8 W5M9K1 A0A087Y256 A0A3B3Z515 A0A2M4B8W8 A0A0K0QRF3 M4AW90 A0A2M4B938 A0A146P5T1 A0A146P740 A0A075IJV4 A0A096M0W0 I3KQM7 A0A1A7Y0I0 A0A3B5PVH2 A0A096M5E1 A0A087XET4 A0A3P9DDV5 A0A1A8C426 A0A3B1J7U9 A0A3B3YP57 A0A1A8JWQ8 A0A3B3YNW5 A0A3B4FJD9 F1QNX5 B7SNG4 A0A1A8QE15 A0A2M3YYU6 A0A1A8L188 A0A1S3R4V3 A0A2M3YYA3 A0A3P8PHL2 G3WLB9 A0A146N073 A0A3B3VRB6 A0A1A7ZXN0 G1T855 G1TLR2 F1PXU6 A0A076FVY1 I3MHA3 A0A384BUF8 A0A2I4AKQ2 A0A3P8W3R3 G1PWB2 U6D754 A0A250Y7G3 G1LGG8 A0A2Y9GBI4 M3WPQ6 A0A2U3ZLZ8 A0A2U9C2U4 A0A091KYJ0
PDB
3LVP
E-value=4.25598e-129,
Score=1185
Ontologies
GO
GO:0046777
GO:0043548
GO:0043560
GO:0016021
GO:0007169
GO:0004714
GO:0005524
GO:0071469
GO:0005887
GO:0042593
GO:0045429
GO:0043559
GO:0048639
GO:0044877
GO:0031995
GO:0005009
GO:0000187
GO:0005635
GO:0006355
GO:0030335
GO:0060267
GO:0051425
GO:0005899
GO:0031981
GO:0005901
GO:0045840
GO:0007186
GO:0005525
GO:0019087
GO:0051290
GO:0003007
GO:0008284
GO:0045821
GO:0045995
GO:0045725
GO:0032148
GO:0001540
GO:0031994
GO:0046326
GO:0005159
GO:0051897
GO:0043235
GO:0043410
GO:0048856
GO:0030424
GO:0014068
GO:0048589
GO:0043009
GO:0005886
GO:0005520
GO:0046328
GO:0005158
GO:0006955
GO:0005010
GO:0048015
GO:0051262
GO:0043066
GO:0043231
GO:0051389
GO:1904646
GO:0042802
GO:0038083
GO:0097242
GO:0035867
GO:0048009
GO:0071333
GO:0005515
GO:0004672
GO:0006468
GO:0016020
GO:0004713
GO:0006281
GO:0048384
PANTHER
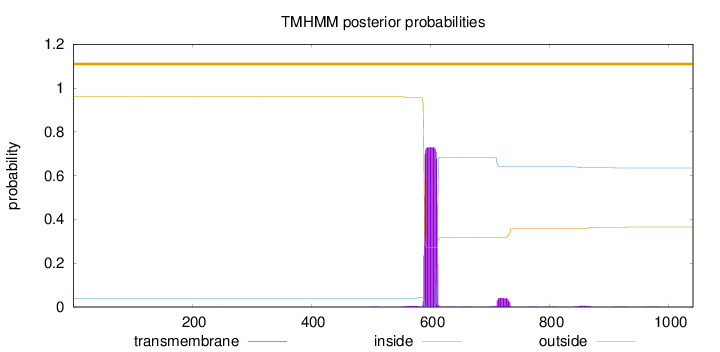
Topology
Length:
1041
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.76509
Exp number, first 60 AAs:
0.00192
Total prob of N-in:
0.03966
outside
1 - 1041
Population Genetic Test Statistics
Pi
253.366281
Theta
186.290042
Tajima's D
1.088956
CLR
0.160238
CSRT
0.686665666716664
Interpretation
Uncertain
