Gene
KWMTBOMO02880 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003656
Annotation
cystathionine_gamma-lyase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.67
Sequence
CDS
ATGGCAGATCACGGATTTCTAAAGCAAAAACCTGGTTTTGCTACTATCGCTATTCACGCTGGCCAAGAACCGGAAAAATGGAATTCAGCAGCCGTGGTTACACCCATTGTCACATCCACTACATTCAAACAGCCAGCACCGGCCGAACATACGGGTTTTGAATACGGTCGCTCTGGAAATCCTACTAGAAACACCCTAGAAGAATGCTTAGCTGCATTAGACGGCGGAAAATACGGTCTAACGTTCGCTTCTGGCCTCGGAGCGACCACAACTTTGGTTGCTCTGCTAAATCACGGAGATCACATCGTGTCCTCAGATGATGTATACGGCGGTACAAACAGACTATTAAGGCAAGTAATCGCAAGATTGGGAATCAACACTACATTTACGGACTTTACAAATATTGAAAAAGTAAAGAACGCCATGCAAGAAAACACCAAGATGATCTGGATAGAGACCCCAACAAATCCATTATTAAAAGTCGTTGACATTGAGGCGATCGTAAAGATCGCGAGGAGTCGCGGAGACATTATCGTTGTTGTAGACAATACCTTCTTGACGCCGTATTTACAAAGACCTTTAGATTTCGGAGCGGACATCGTTATGTACTCGCTAACGAAATATATGAATGGTCATTCTGACGTCATAATGGGAGCCGCTATTGTAAATGACAAAGCTATCGGAGACAAACTAAGATTCTTGCAAAACGCCATGGGCATCGTTCCGTCACCGATGGACTGCTATTTTGTGAACAGGAGCTTAAAGACGTTAGCATTAAGAATGGAGCATCACAAGAAGTCGTCATTGATCATTGCCAATTGGTTGGAGAAACATCCGAAAATCGTCGAAGTGCTTCACCCAGGGCTGCCAACTCACCCGCAACACGAGATAGCTAAAAAACAGATGACGGGTCACTCGGGTGTGTTCAGTTTCAAACATTCCGGAGGTCTCAAGGAATCTAGAAAATTCCTGAGCTCATTGAAAGTGTTCACGCTGGCCGAGAGCTTGGGGGGCTATGAAAGTCTCGCTGAGCTACCATCTGTGATGACACACGCATCAGTTCCGGCGCTGCAGAGGGAGGAACTCGGCATCACCGATTCGCTGGTAAGACTTTCCGTCGGTCTCGAAGATACCGAGGATCTTATCGAAGATTTAGATCAAGCTTTTAAAGCTGCATTTTAA
Protein
MADHGFLKQKPGFATIAIHAGQEPEKWNSAAVVTPIVTSTTFKQPAPAEHTGFEYGRSGNPTRNTLEECLAALDGGKYGLTFASGLGATTTLVALLNHGDHIVSSDDVYGGTNRLLRQVIARLGINTTFTDFTNIEKVKNAMQENTKMIWIETPTNPLLKVVDIEAIVKIARSRGDIIVVVDNTFLTPYLQRPLDFGADIVMYSLTKYMNGHSDVIMGAAIVNDKAIGDKLRFLQNAMGIVPSPMDCYFVNRSLKTLALRMEHHKKSSLIIANWLEKHPKIVEVLHPGLPTHPQHEIAKKQMTGHSGVFSFKHSGGLKESRKFLSSLKVFTLAESLGGYESLAELPSVMTHASVPALQREELGITDSLVRLSVGLEDTEDLIEDLDQAFKAAF
Summary
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the trans-sulfuration enzymes family.
Uniprot
Q2F6B7
H9J2B9
A0A0L7LMD3
A0A194QUM6
A0A194PNV8
A0A2W1BMT7
+ More
A0A2H1VG61 A0A2A4IWX7 A0A212F2K2 A0A2W1BSU2 A0A2A4K9Q2 A0A1Q3F233 A0A182HWD8 Q7QHF8 A0A182X5L1 A0A182V506 A0A182JNJ4 B0W7Q2 A0A2M4CUD7 A0A182U5L0 Q17DR0 A0A1S4F6U1 A0A2M4CUB2 A0A2M4BRB6 A0A182NZK9 A0A182M218 A0A182Q2Z1 A0A2M3Z4L3 A0A182FKJ4 A0A182RTE3 A0A023ETT3 A0A182YB36 T1DJR2 A0A182JKI4 A0A2M4BQL6 A0A182H2J4 A0A182N2C9 A0A182WDH5 A0A2M4AND2 A0A182S739 A0A2H1V2G9 A0A2M4AMA8 A0A2H1V295 A0A1A9WFW0 A0A0L0CDY9 A0A0K8TTP1 U5EJE2 A0A1I8MHH1 A0A2J7QCJ1 A0A1I8QAD5 U4U6P1 V5SIY2 N6UBY9 A0A1B0AMY2 A0A1Y1JWR5 K1R7F6 A0A1L8EC44 E9IFN6 B4MG50 B3MDE6 B4QD03 A0A1A9VUQ6 B4KJJ5 E9HXC9 B3NK88 B4HNZ0 B4JPP3 A0A0P5YPG7 A0A0A1X101 A0A0P5DQG7 D3TMY0 A0A034VR89 A0A2H1V296 B4P4B7 A0A0K8VR58 W8B0N5 B4MP07 A0A0P6HL73 A0A0P5HYY7 Q7JXZ2 A0A3B0KBQ4 T1JND1 A0A232FMQ8 K7IVZ2 W5J4H5 O97121 B4M0Z9 A0A1L8E2A6 A0A0P6B4N4 V4A6N7 A0A1W4VQN3 A0A336LSS6 A0A2P8ZKX8 E0VK61 A0A1J1I6S3 A0A084VHV3 A0A0P4ZWP4 A0A026WE10 A0A1L8E9P3 A0A1S3JF62
A0A2H1VG61 A0A2A4IWX7 A0A212F2K2 A0A2W1BSU2 A0A2A4K9Q2 A0A1Q3F233 A0A182HWD8 Q7QHF8 A0A182X5L1 A0A182V506 A0A182JNJ4 B0W7Q2 A0A2M4CUD7 A0A182U5L0 Q17DR0 A0A1S4F6U1 A0A2M4CUB2 A0A2M4BRB6 A0A182NZK9 A0A182M218 A0A182Q2Z1 A0A2M3Z4L3 A0A182FKJ4 A0A182RTE3 A0A023ETT3 A0A182YB36 T1DJR2 A0A182JKI4 A0A2M4BQL6 A0A182H2J4 A0A182N2C9 A0A182WDH5 A0A2M4AND2 A0A182S739 A0A2H1V2G9 A0A2M4AMA8 A0A2H1V295 A0A1A9WFW0 A0A0L0CDY9 A0A0K8TTP1 U5EJE2 A0A1I8MHH1 A0A2J7QCJ1 A0A1I8QAD5 U4U6P1 V5SIY2 N6UBY9 A0A1B0AMY2 A0A1Y1JWR5 K1R7F6 A0A1L8EC44 E9IFN6 B4MG50 B3MDE6 B4QD03 A0A1A9VUQ6 B4KJJ5 E9HXC9 B3NK88 B4HNZ0 B4JPP3 A0A0P5YPG7 A0A0A1X101 A0A0P5DQG7 D3TMY0 A0A034VR89 A0A2H1V296 B4P4B7 A0A0K8VR58 W8B0N5 B4MP07 A0A0P6HL73 A0A0P5HYY7 Q7JXZ2 A0A3B0KBQ4 T1JND1 A0A232FMQ8 K7IVZ2 W5J4H5 O97121 B4M0Z9 A0A1L8E2A6 A0A0P6B4N4 V4A6N7 A0A1W4VQN3 A0A336LSS6 A0A2P8ZKX8 E0VK61 A0A1J1I6S3 A0A084VHV3 A0A0P4ZWP4 A0A026WE10 A0A1L8E9P3 A0A1S3JF62
Pubmed
19121390
26227816
26354079
28756777
22118469
12364791
+ More
14747013 17210077 17510324 24945155 25244985 26483478 26108605 26369729 25315136 23537049 28004739 22992520 21282665 17994087 22936249 21292972 25830018 20353571 25348373 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20075255 20920257 23761445 10319443 23254933 29403074 20566863 24438588 24508170
14747013 17210077 17510324 24945155 25244985 26483478 26108605 26369729 25315136 23537049 28004739 22992520 21282665 17994087 22936249 21292972 25830018 20353571 25348373 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20075255 20920257 23761445 10319443 23254933 29403074 20566863 24438588 24508170
EMBL
DQ311155
ABD36100.1
BABH01007644
BABH01007645
BABH01007646
JTDY01000577
+ More
KOB76602.1 KQ461150 KPJ08680.1 KQ459597 KPI94997.1 KZ149960 PZC76382.1 ODYU01002386 SOQ39817.1 NWSH01006069 PCG63692.1 AGBW02010734 OWR47951.1 KZ150027 PZC74793.1 NWSH01000034 PCG80460.1 GFDL01013444 JAV21601.1 APCN01001485 AAAB01008816 EAA05138.3 DS231855 EDS38196.1 GGFL01004755 MBW68933.1 CH477291 EAT44603.1 GGFL01004756 MBW68934.1 GGFJ01006382 MBW55523.1 AXCM01000599 AXCN02001727 GGFM01002692 MBW23443.1 GAPW01001769 JAC11829.1 GAMD01001316 JAB00275.1 GGFJ01006235 MBW55376.1 JXUM01105481 JXUM01105482 KQ564969 KXJ71493.1 GGFK01008974 MBW42295.1 ODYU01000352 SOQ34966.1 GGFK01008602 MBW41923.1 SOQ34965.1 JRES01000520 KNC30451.1 GDAI01000085 JAI17518.1 GANO01002264 JAB57607.1 NEVH01016289 PNF26293.1 KB631805 ERL86281.1 KF647650 AHB50512.1 APGK01029504 KB740735 ENN79170.1 JXJN01000640 GEZM01098785 JAV53759.1 JH816312 EKC29931.1 GFDG01002594 JAV16205.1 GL762880 EFZ20610.1 CH940671 EDW58188.1 CH902619 EDV36394.1 CM000362 CM002911 EDX07708.1 KMY94895.1 CH933807 EDW13575.2 GL733029 EFX63600.1 CH954179 EDV55110.1 CH480816 EDW48492.1 CH916372 EDV98873.1 GDIP01246284 GDIP01068900 LRGB01002864 JAM34815.1 KZS05728.1 GBXI01009520 JAD04772.1 GDIP01152898 JAJ70504.1 EZ422782 ADD19058.1 GAKP01014330 JAC44622.1 SOQ34967.1 CM000158 EDW91603.1 GDHF01010952 JAI41362.1 GAMC01011900 JAB94655.1 CH963848 EDW73846.1 GDIQ01025914 JAN68823.1 GDIQ01226758 JAK24967.1 AE013599 AY089545 AAF57663.1 AAL90283.1 AHN56373.1 OUUW01000010 SPP85600.1 JH431789 NNAY01000040 OXU31647.1 ADMH02002090 ETN59372.1 AF095822 AAD17839.1 CH940650 EDW67410.1 GFDF01001214 JAV12870.1 GDIP01022176 JAM81539.1 KB202094 ESO92362.1 UFQT01000155 SSX20980.1 PYGN01000026 PSN57157.1 DS235239 EEB13767.1 CVRI01000041 CRK95268.1 ATLV01013237 KE524847 KFB37547.1 GDIP01211341 JAJ12061.1 KK107262 EZA53916.1 GFDG01003352 JAV15447.1
KOB76602.1 KQ461150 KPJ08680.1 KQ459597 KPI94997.1 KZ149960 PZC76382.1 ODYU01002386 SOQ39817.1 NWSH01006069 PCG63692.1 AGBW02010734 OWR47951.1 KZ150027 PZC74793.1 NWSH01000034 PCG80460.1 GFDL01013444 JAV21601.1 APCN01001485 AAAB01008816 EAA05138.3 DS231855 EDS38196.1 GGFL01004755 MBW68933.1 CH477291 EAT44603.1 GGFL01004756 MBW68934.1 GGFJ01006382 MBW55523.1 AXCM01000599 AXCN02001727 GGFM01002692 MBW23443.1 GAPW01001769 JAC11829.1 GAMD01001316 JAB00275.1 GGFJ01006235 MBW55376.1 JXUM01105481 JXUM01105482 KQ564969 KXJ71493.1 GGFK01008974 MBW42295.1 ODYU01000352 SOQ34966.1 GGFK01008602 MBW41923.1 SOQ34965.1 JRES01000520 KNC30451.1 GDAI01000085 JAI17518.1 GANO01002264 JAB57607.1 NEVH01016289 PNF26293.1 KB631805 ERL86281.1 KF647650 AHB50512.1 APGK01029504 KB740735 ENN79170.1 JXJN01000640 GEZM01098785 JAV53759.1 JH816312 EKC29931.1 GFDG01002594 JAV16205.1 GL762880 EFZ20610.1 CH940671 EDW58188.1 CH902619 EDV36394.1 CM000362 CM002911 EDX07708.1 KMY94895.1 CH933807 EDW13575.2 GL733029 EFX63600.1 CH954179 EDV55110.1 CH480816 EDW48492.1 CH916372 EDV98873.1 GDIP01246284 GDIP01068900 LRGB01002864 JAM34815.1 KZS05728.1 GBXI01009520 JAD04772.1 GDIP01152898 JAJ70504.1 EZ422782 ADD19058.1 GAKP01014330 JAC44622.1 SOQ34967.1 CM000158 EDW91603.1 GDHF01010952 JAI41362.1 GAMC01011900 JAB94655.1 CH963848 EDW73846.1 GDIQ01025914 JAN68823.1 GDIQ01226758 JAK24967.1 AE013599 AY089545 AAF57663.1 AAL90283.1 AHN56373.1 OUUW01000010 SPP85600.1 JH431789 NNAY01000040 OXU31647.1 ADMH02002090 ETN59372.1 AF095822 AAD17839.1 CH940650 EDW67410.1 GFDF01001214 JAV12870.1 GDIP01022176 JAM81539.1 KB202094 ESO92362.1 UFQT01000155 SSX20980.1 PYGN01000026 PSN57157.1 DS235239 EEB13767.1 CVRI01000041 CRK95268.1 ATLV01013237 KE524847 KFB37547.1 GDIP01211341 JAJ12061.1 KK107262 EZA53916.1 GFDG01003352 JAV15447.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000075840 UP000007062 UP000076407 UP000075903 UP000075881 UP000002320 UP000075902 UP000008820 UP000075885 UP000075883 UP000075886 UP000069272 UP000075900 UP000076408 UP000075880 UP000069940 UP000249989 UP000075884 UP000075920 UP000075901 UP000091820 UP000037069 UP000095301 UP000235965 UP000095300 UP000030742 UP000019118 UP000092460 UP000005408 UP000008792 UP000007801 UP000000304 UP000078200 UP000009192 UP000000305 UP000008711 UP000001292 UP000001070 UP000076858 UP000002282 UP000007798 UP000000803 UP000268350 UP000215335 UP000002358 UP000000673 UP000030746 UP000192221 UP000245037 UP000009046 UP000183832 UP000030765 UP000053097 UP000085678
UP000075840 UP000007062 UP000076407 UP000075903 UP000075881 UP000002320 UP000075902 UP000008820 UP000075885 UP000075883 UP000075886 UP000069272 UP000075900 UP000076408 UP000075880 UP000069940 UP000249989 UP000075884 UP000075920 UP000075901 UP000091820 UP000037069 UP000095301 UP000235965 UP000095300 UP000030742 UP000019118 UP000092460 UP000005408 UP000008792 UP000007801 UP000000304 UP000078200 UP000009192 UP000000305 UP000008711 UP000001292 UP000001070 UP000076858 UP000002282 UP000007798 UP000000803 UP000268350 UP000215335 UP000002358 UP000000673 UP000030746 UP000192221 UP000245037 UP000009046 UP000183832 UP000030765 UP000053097 UP000085678
Pfam
PF01053 Cys_Met_Meta_PP
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
CDD
ProteinModelPortal
Q2F6B7
H9J2B9
A0A0L7LMD3
A0A194QUM6
A0A194PNV8
A0A2W1BMT7
+ More
A0A2H1VG61 A0A2A4IWX7 A0A212F2K2 A0A2W1BSU2 A0A2A4K9Q2 A0A1Q3F233 A0A182HWD8 Q7QHF8 A0A182X5L1 A0A182V506 A0A182JNJ4 B0W7Q2 A0A2M4CUD7 A0A182U5L0 Q17DR0 A0A1S4F6U1 A0A2M4CUB2 A0A2M4BRB6 A0A182NZK9 A0A182M218 A0A182Q2Z1 A0A2M3Z4L3 A0A182FKJ4 A0A182RTE3 A0A023ETT3 A0A182YB36 T1DJR2 A0A182JKI4 A0A2M4BQL6 A0A182H2J4 A0A182N2C9 A0A182WDH5 A0A2M4AND2 A0A182S739 A0A2H1V2G9 A0A2M4AMA8 A0A2H1V295 A0A1A9WFW0 A0A0L0CDY9 A0A0K8TTP1 U5EJE2 A0A1I8MHH1 A0A2J7QCJ1 A0A1I8QAD5 U4U6P1 V5SIY2 N6UBY9 A0A1B0AMY2 A0A1Y1JWR5 K1R7F6 A0A1L8EC44 E9IFN6 B4MG50 B3MDE6 B4QD03 A0A1A9VUQ6 B4KJJ5 E9HXC9 B3NK88 B4HNZ0 B4JPP3 A0A0P5YPG7 A0A0A1X101 A0A0P5DQG7 D3TMY0 A0A034VR89 A0A2H1V296 B4P4B7 A0A0K8VR58 W8B0N5 B4MP07 A0A0P6HL73 A0A0P5HYY7 Q7JXZ2 A0A3B0KBQ4 T1JND1 A0A232FMQ8 K7IVZ2 W5J4H5 O97121 B4M0Z9 A0A1L8E2A6 A0A0P6B4N4 V4A6N7 A0A1W4VQN3 A0A336LSS6 A0A2P8ZKX8 E0VK61 A0A1J1I6S3 A0A084VHV3 A0A0P4ZWP4 A0A026WE10 A0A1L8E9P3 A0A1S3JF62
A0A2H1VG61 A0A2A4IWX7 A0A212F2K2 A0A2W1BSU2 A0A2A4K9Q2 A0A1Q3F233 A0A182HWD8 Q7QHF8 A0A182X5L1 A0A182V506 A0A182JNJ4 B0W7Q2 A0A2M4CUD7 A0A182U5L0 Q17DR0 A0A1S4F6U1 A0A2M4CUB2 A0A2M4BRB6 A0A182NZK9 A0A182M218 A0A182Q2Z1 A0A2M3Z4L3 A0A182FKJ4 A0A182RTE3 A0A023ETT3 A0A182YB36 T1DJR2 A0A182JKI4 A0A2M4BQL6 A0A182H2J4 A0A182N2C9 A0A182WDH5 A0A2M4AND2 A0A182S739 A0A2H1V2G9 A0A2M4AMA8 A0A2H1V295 A0A1A9WFW0 A0A0L0CDY9 A0A0K8TTP1 U5EJE2 A0A1I8MHH1 A0A2J7QCJ1 A0A1I8QAD5 U4U6P1 V5SIY2 N6UBY9 A0A1B0AMY2 A0A1Y1JWR5 K1R7F6 A0A1L8EC44 E9IFN6 B4MG50 B3MDE6 B4QD03 A0A1A9VUQ6 B4KJJ5 E9HXC9 B3NK88 B4HNZ0 B4JPP3 A0A0P5YPG7 A0A0A1X101 A0A0P5DQG7 D3TMY0 A0A034VR89 A0A2H1V296 B4P4B7 A0A0K8VR58 W8B0N5 B4MP07 A0A0P6HL73 A0A0P5HYY7 Q7JXZ2 A0A3B0KBQ4 T1JND1 A0A232FMQ8 K7IVZ2 W5J4H5 O97121 B4M0Z9 A0A1L8E2A6 A0A0P6B4N4 V4A6N7 A0A1W4VQN3 A0A336LSS6 A0A2P8ZKX8 E0VK61 A0A1J1I6S3 A0A084VHV3 A0A0P4ZWP4 A0A026WE10 A0A1L8E9P3 A0A1S3JF62
PDB
3COG
E-value=3.9948e-126,
Score=1156
Ontologies
PATHWAY
00260
Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00450 Selenocompound metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00450 Selenocompound metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
PANTHER
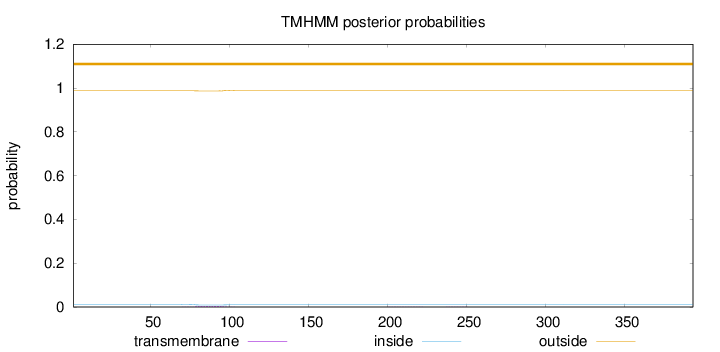
Topology
Length:
393
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0893699999999999
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.01232
outside
1 - 393
Population Genetic Test Statistics
Pi
280.512334
Theta
212.602225
Tajima's D
0.827776
CLR
0.389002
CSRT
0.610269486525674
Interpretation
Uncertain
