Gene
KWMTBOMO02875
Annotation
PREDICTED:_cytosolic_carboxypeptidase_4-like_[Bombyx_mori]
Full name
Cytosolic carboxypeptidase 1
Alternative Name
ATP/GTP-binding protein 1
Location in the cell
Extracellular Reliability : 1.087 Nuclear Reliability : 1.444
Sequence
CDS
ATGTTGCCAGCCTTGATGCACAGAATATCACCTGCTTTTGCACTCGGTTCGTGTTCTTTCCGTGTAGAAAGAGAAAGGGAGAGTACCGCTCGGGTAACGGTATGGAGACACCTTGGTGTTACGCGCTCTTACACAATGGAAGCCTCTTTCTGCGGTTTCGATCGTGGACCTTTTAAGGGTTTCCACCTGAACACCCAGCACCTGCAGAATGTCGGCAGCGACTTTTGTGAAGCTCTTAATGGATTGAACGACAGCGCTGCTAATGTCGACATTCAGCTAACGAAAGACCTGAATGGCGAGGCAGCGGTGGAAAGCGAACTGGGCTCCGGCTCTGATAGCGTTCTGAAAACCGATTCAGATGAAGACTTTGATTAG
Protein
MLPALMHRISPAFALGSCSFRVERERESTARVTVWRHLGVTRSYTMEASFCGFDRGPFKGFHLNTQHLQNVGSDFCEALNGLNDSAANVDIQLTKDLNGEAAVESELGSGSDSVLKTDSDEDFD
Summary
Description
Metallocarboxypeptidase that mediates deglutamylation of target proteins. Catalyzes the deglutamylation of polyglutamate side chains generated by post-translational polyglutamylation in proteins such as tubulins. Also removes gene-encoded polyglutamates from the carboxy-terminus of target proteins. Acts as a long-chain deglutamylase and specifically shortens long polyglutamate chains, while it is not able to remove the branching point glutamate.
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M14 family.
Keywords
Carboxypeptidase
Complete proteome
Cytoplasm
Hydrolase
Metal-binding
Metalloprotease
Mitochondrion
Nucleus
Protease
Reference proteome
Zinc
Feature
chain Cytosolic carboxypeptidase 1
Uniprot
A0A2W1BVR6
A0A212FLY7
A0A2H1X2J4
A0A2A4K1G4
A0A2A4K2V9
A0A194QYM5
+ More
A0A194PPU5 A0A0L7LG24 A0A0C9QI19 A0A0L7RBK9 A0A2A3EUK7 A0A154PLW7 A0A310SG35 A0A088ACX1 A0A3L8DG93 A0A195BPK7 A0A091NAW7 A0A093FAN0 A0A091MH26 A0A094L4A4 U3IVU6 A0A2P4SWC8 H0YXY9 A0A091MZ86 Q80TL9 A0A1S3JTQ9 A0A226NB39 A0A226PKT4 A0A3P8W1W9 A0A2D4JYU8 A0A2H6N9Y7 A0A286YDV2 A0A3P8VZ54 A0A1B6K6N5 G3NXR8 E1C3P4 A0A1S3JTZ6 A0A1S3JTA4 R9PXP2 U3JLV1 G1N3X3 G1N3Y3 A0A2C9KMQ5 A0A2I0UGW3 A0A2C9KMW1 A0A3P9LD14 A0A3P9HHF1 A0A2C9KMQ7 A0A3P9J2P8 H2LKZ5 A0A218V952 H2LKZ4 H2LKZ6 A0A091TTM2 A0A3P9LD56
A0A194PPU5 A0A0L7LG24 A0A0C9QI19 A0A0L7RBK9 A0A2A3EUK7 A0A154PLW7 A0A310SG35 A0A088ACX1 A0A3L8DG93 A0A195BPK7 A0A091NAW7 A0A093FAN0 A0A091MH26 A0A094L4A4 U3IVU6 A0A2P4SWC8 H0YXY9 A0A091MZ86 Q80TL9 A0A1S3JTQ9 A0A226NB39 A0A226PKT4 A0A3P8W1W9 A0A2D4JYU8 A0A2H6N9Y7 A0A286YDV2 A0A3P8VZ54 A0A1B6K6N5 G3NXR8 E1C3P4 A0A1S3JTZ6 A0A1S3JTA4 R9PXP2 U3JLV1 G1N3X3 G1N3Y3 A0A2C9KMQ5 A0A2I0UGW3 A0A2C9KMW1 A0A3P9LD14 A0A3P9HHF1 A0A2C9KMQ7 A0A3P9J2P8 H2LKZ5 A0A218V952 H2LKZ4 H2LKZ6 A0A091TTM2 A0A3P9LD56
EC Number
3.4.17.-
Pubmed
EMBL
KZ149903
PZC78431.1
AGBW02007682
OWR54763.1
ODYU01012974
SOQ59543.1
+ More
NWSH01000238 PCG78111.1 PCG78113.1 KQ461150 KPJ08686.1 KQ459597 KPI94993.1 JTDY01001263 KOB74350.1 GBYB01014338 JAG84105.1 KQ414617 KOC68249.1 KZ288185 PBC34962.1 KQ434977 KZC12842.1 KQ773785 OAD52362.1 QOIP01000008 RLU19331.1 KQ976424 KYM88452.1 KL379274 KFP86177.1 KK390971 KFV54722.1 KK825849 KFP72695.1 KL359261 KFZ64322.1 PPHD01019402 POI28421.1 ABQF01033238 ABQF01033239 KK840161 KFP82808.1 AK122423 BAC65705.1 MCFN01000111 OXB64804.1 AWGT02000056 OXB80122.1 IACL01025140 LAB01574.1 IACI01077717 LAA27867.1 AC135469 AC155298 GECU01036341 GECU01001020 GECU01000594 JAS71365.1 JAT06687.1 JAT07113.1 AADN02064844 AADN02064845 AADN02064846 AADN02064847 AADN02064848 AADN02064849 AADN02064850 AADN02064851 AC186856 AGTO01001391 AGTO01021630 KZ505770 PKU45270.1 MUZQ01000025 OWK62554.1 KK486085 KFQ62006.1
NWSH01000238 PCG78111.1 PCG78113.1 KQ461150 KPJ08686.1 KQ459597 KPI94993.1 JTDY01001263 KOB74350.1 GBYB01014338 JAG84105.1 KQ414617 KOC68249.1 KZ288185 PBC34962.1 KQ434977 KZC12842.1 KQ773785 OAD52362.1 QOIP01000008 RLU19331.1 KQ976424 KYM88452.1 KL379274 KFP86177.1 KK390971 KFV54722.1 KK825849 KFP72695.1 KL359261 KFZ64322.1 PPHD01019402 POI28421.1 ABQF01033238 ABQF01033239 KK840161 KFP82808.1 AK122423 BAC65705.1 MCFN01000111 OXB64804.1 AWGT02000056 OXB80122.1 IACL01025140 LAB01574.1 IACI01077717 LAA27867.1 AC135469 AC155298 GECU01036341 GECU01001020 GECU01000594 JAS71365.1 JAT06687.1 JAT07113.1 AADN02064844 AADN02064845 AADN02064846 AADN02064847 AADN02064848 AADN02064849 AADN02064850 AADN02064851 AC186856 AGTO01001391 AGTO01021630 KZ505770 PKU45270.1 MUZQ01000025 OWK62554.1 KK486085 KFQ62006.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000053268
UP000037510
UP000053825
+ More
UP000242457 UP000076502 UP000005203 UP000279307 UP000078540 UP000007754 UP000085678 UP000198323 UP000198419 UP000265120 UP000000589 UP000007635 UP000000539 UP000016665 UP000001645 UP000076420 UP000265180 UP000265200 UP000001038 UP000197619
UP000242457 UP000076502 UP000005203 UP000279307 UP000078540 UP000007754 UP000085678 UP000198323 UP000198419 UP000265120 UP000000589 UP000007635 UP000000539 UP000016665 UP000001645 UP000076420 UP000265180 UP000265200 UP000001038 UP000197619
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
CDD
ProteinModelPortal
A0A2W1BVR6
A0A212FLY7
A0A2H1X2J4
A0A2A4K1G4
A0A2A4K2V9
A0A194QYM5
+ More
A0A194PPU5 A0A0L7LG24 A0A0C9QI19 A0A0L7RBK9 A0A2A3EUK7 A0A154PLW7 A0A310SG35 A0A088ACX1 A0A3L8DG93 A0A195BPK7 A0A091NAW7 A0A093FAN0 A0A091MH26 A0A094L4A4 U3IVU6 A0A2P4SWC8 H0YXY9 A0A091MZ86 Q80TL9 A0A1S3JTQ9 A0A226NB39 A0A226PKT4 A0A3P8W1W9 A0A2D4JYU8 A0A2H6N9Y7 A0A286YDV2 A0A3P8VZ54 A0A1B6K6N5 G3NXR8 E1C3P4 A0A1S3JTZ6 A0A1S3JTA4 R9PXP2 U3JLV1 G1N3X3 G1N3Y3 A0A2C9KMQ5 A0A2I0UGW3 A0A2C9KMW1 A0A3P9LD14 A0A3P9HHF1 A0A2C9KMQ7 A0A3P9J2P8 H2LKZ5 A0A218V952 H2LKZ4 H2LKZ6 A0A091TTM2 A0A3P9LD56
A0A194PPU5 A0A0L7LG24 A0A0C9QI19 A0A0L7RBK9 A0A2A3EUK7 A0A154PLW7 A0A310SG35 A0A088ACX1 A0A3L8DG93 A0A195BPK7 A0A091NAW7 A0A093FAN0 A0A091MH26 A0A094L4A4 U3IVU6 A0A2P4SWC8 H0YXY9 A0A091MZ86 Q80TL9 A0A1S3JTQ9 A0A226NB39 A0A226PKT4 A0A3P8W1W9 A0A2D4JYU8 A0A2H6N9Y7 A0A286YDV2 A0A3P8VZ54 A0A1B6K6N5 G3NXR8 E1C3P4 A0A1S3JTZ6 A0A1S3JTA4 R9PXP2 U3JLV1 G1N3X3 G1N3Y3 A0A2C9KMQ5 A0A2I0UGW3 A0A2C9KMW1 A0A3P9LD14 A0A3P9HHF1 A0A2C9KMQ7 A0A3P9J2P8 H2LKZ5 A0A218V952 H2LKZ4 H2LKZ6 A0A091TTM2 A0A3P9LD56
Ontologies
GO
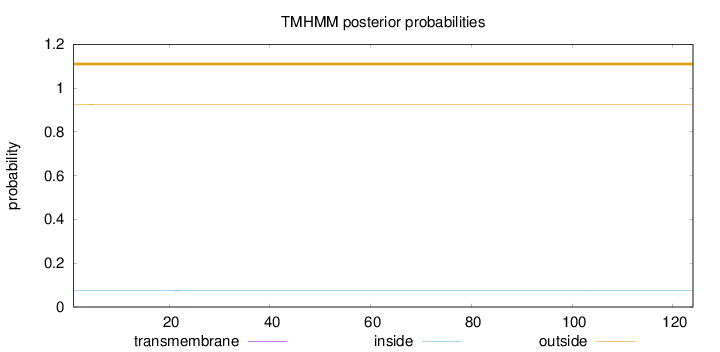
Topology
Subcellular location
Cytoplasm
Cytosol
Nucleus
Mitochondrion
Cytosol
Nucleus
Mitochondrion
Length:
124
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01505
Exp number, first 60 AAs:
0.01505
Total prob of N-in:
0.07375
outside
1 - 124
Population Genetic Test Statistics
Pi
272.582285
Theta
196.226926
Tajima's D
1.910352
CLR
0.198023
CSRT
0.867306634668267
Interpretation
Uncertain
