Gene
KWMTBOMO02873 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003660
Annotation
PREDICTED:_uncharacterized_protein_LOC106140166_isoform_X1_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.455
Sequence
CDS
ATGCAGTTCTTCTTGACATCATGTCTCGTGCTGCTCGCAGCTTCCCTCGGCCCAGCCGCAGCTCAAAGAATAACAACGATACAACTGGATGGCGTTCAGTATTTTATCTCTAGGATGAATCCGTACAGCCCAGAGCTCAACTATTTCCTGTCCTATCAGTACTGCAGATCATTAGGTCTTCAACTGGCATCGTTCGAGACGAAGGAGAAGGCAGATTCGATAACGACTTATCTAACAAATGCAGGTTACAACAAATATGACTTCTGGACATCGGGTAATAACCTCGGCACAGACATGTTTTTGTGGATGAGCACCGGTCTGCCATTTAATGCGACTTTTAACTACATGCGGAGACTGCCCATCGATGCTCCTGCCCAGCATGCTGATGACAGCATGGACCCTCTAGACGTACCACAAGGAAGCACTGCTCCACAACGCACCGCCCGACATGGGACGGAACACGTCATGACTAATGGCTGCATCGCGCTCAAAGCTCCGACCTTCCACTGGGAGCCTCAACACTGCGGCGAGATCAAGGACTTCATATGCGAGCAGACGCGTTGCTATTACTACAACTACGGCTCGATTCCCGTTTCATCCGCGCAGGGAAAGCCGCTATCGCTGACGACGACGACGACCGCCCACCCAATCACGTCGTCCTCGCCACCGCCACCGCGCTCCGAGCACCTGCCGACCCACTTCAACCTTAACAATCTCATCGGCAAGCTGCGACCTTCTTCCCTTGACGGCCTCCAGTCTGCACACCTCAAGACTAGCGCCTTGTTCAAATCCCCACCGCAAATCGACTCCCACTATTCGCGAGCTGCGGACGCTCATTCCCGCGATCACGACCAGGACCAGAAGGAAGAACCCACTCAGGGTCCGTACGAAGATGATGAGGGTATGGTGGGAGACGATCCGGCCGCGTATCTGACTAACGAGGCTCACGACGCTCCCGTTGGAGACGAGATCGCATCGACTGCCGAGCCGGACGCCACCGAGCACTCCGTCCACGCCAGCGGCATGTTGGCGCCCCCCGCTTATTAA
Protein
MQFFLTSCLVLLAASLGPAAAQRITTIQLDGVQYFISRMNPYSPELNYFLSYQYCRSLGLQLASFETKEKADSITTYLTNAGYNKYDFWTSGNNLGTDMFLWMSTGLPFNATFNYMRRLPIDAPAQHADDSMDPLDVPQGSTAPQRTARHGTEHVMTNGCIALKAPTFHWEPQHCGEIKDFICEQTRCYYYNYGSIPVSSAQGKPLSLTTTTTAHPITSSSPPPPRSEHLPTHFNLNNLIGKLRPSSLDGLQSAHLKTSALFKSPPQIDSHYSRAADAHSRDHDQDQKEEPTQGPYEDDEGMVGDDPAAYLTNEAHDAPVGDEIASTAEPDATEHSVHASGMLAPPAY
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00059 Lectin_C
Interpro
SUPFAM
SSF56436
SSF56436
Gene 3D
ProteinModelPortal
Ontologies
GO
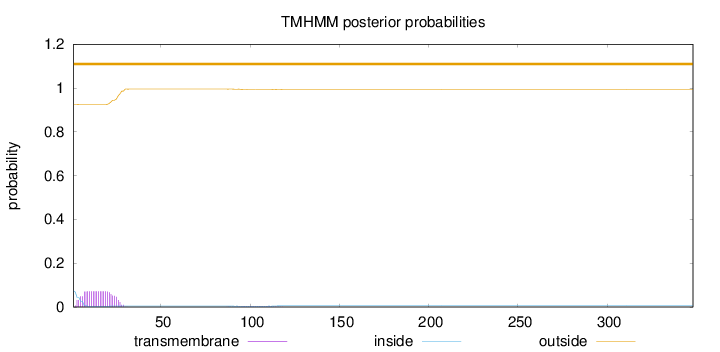
Topology
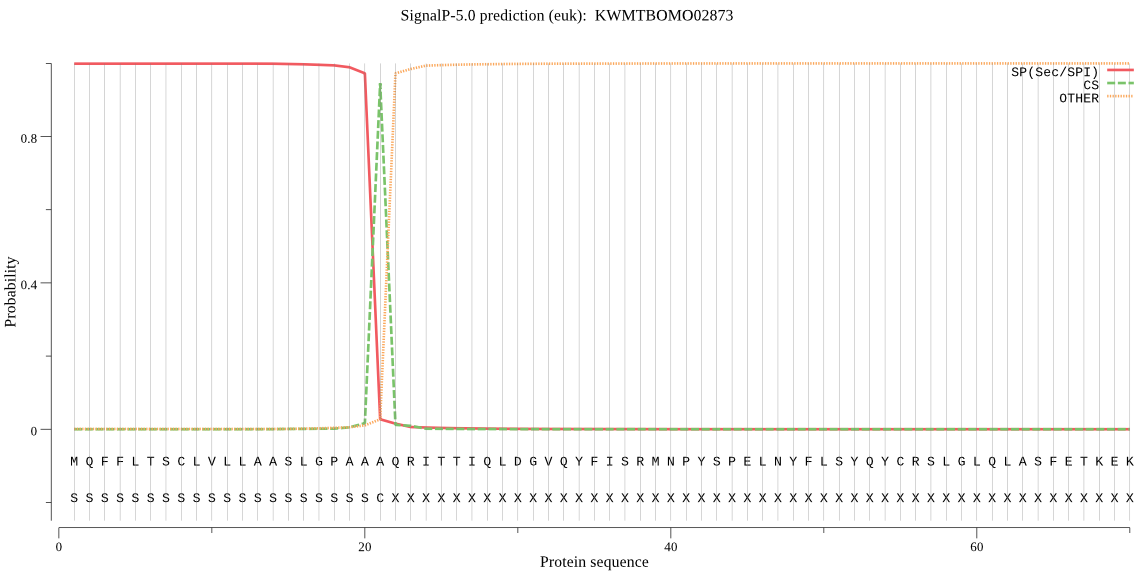
SignalP
Position: 1 - 21,
Likelihood: 0.999138
Length:
348
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.57902
Exp number, first 60 AAs:
1.50993
Total prob of N-in:
0.07457
outside
1 - 348
Population Genetic Test Statistics
Pi
317.456072
Theta
199.297234
Tajima's D
1.572995
CLR
0.323066
CSRT
0.804659767011649
Interpretation
Uncertain
