Gene
KWMTBOMO02867
Annotation
PREDICTED:_uncharacterized_protein_LOC105842183_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.397
Sequence
CDS
ATGAACTTTGTTTTCATAATACTGATTTTAGGATGGACATTTATGACTACTGTACAGGACTGTCCTGAAGAAATTTCGCATAGTGGAATTAGCGTCAAATGGAAACAAAGTTCTTGGGCTAATCATATAGAATCTGATCCGCAATGCTACCGAAACCAAAATCTAATCACAAGAAACTGTACAGATAACGTTTGGAAGCCGCCATTAGAAGCTGTCGAACCTTGTCTCAAGGTCGTAAAGAATTTTGACGTATTTTCTTGTCCTCCTGGATATCACAAAATATCAGAAAAGAACAATGACTATTGTTATCAAATTGGCCGACCATCACAATGGGAATATCCTTGCTTTAGCAGTGGAGCGGCATCGGTCATCACAGATTTATCAGAACTCGAATTAAATTCTTTACTGCAGTCCCTGGTTTCAATGAATATATCTAGATATTTTTGGCTACCCGGAAAAAGACACAAACTTTTTAATCCAGCTGTTTGGAACATCCCAGGTCCAAACTGGGGTCGACGTGTGAAGTCTAACAGCTACTTGCGTTACAGGACGATATTTTTAAAAAATTGCCTCTTACTTGATGTTCAAAAGCGAGTGATCATCACAGAAACCTGTAGTAACATTTACCCTAGTTTGTGTTTCTATATTAACGAATTTTATCATCCGTCTAAATGTCCTGACGGATATTATGCACTTAGATTTCAACTTGATAACGGCACTTGCTTCGGAATTGAAAGCTCAGATTCGATAAATGGTTGGACTTTTGCCGAATTCATAACCGCCAAATGTAAAAATCCAATGAACGTTTCTTCATTGAATGCTTTGAGTAGATTCATGTTCAAAAAAATTGCTGAACTTTCTAAATTACCAAACGAAATTTGGTGCTGGTTAAAATTTACCAGTTACTATATGGCTAGCAAAAATAATACTTTCTTGGAAGACTTAGAAGAACATAATTTAGAAGGTATAATAGATAATGCCGGAACTTTAGGATTAATGAATAAATCACGGCACCTGTCTTGTATGGCATGTGAAACTCAAGTTGTTTATTTAGAAACAGAGTTGATGTTTGAATATAATACAGTCGATAACAATCTTTACCTCACTATATATTATCCTTCAGGTTTATGGAAATATAATAAGACCGATAAAGGTGTTCAGTGTTTTTCCGATGCCAATGGATTTGTGGGTGTCATAAATGTACGTGAACTTGATTTATTTCAATCAAAACAAATAATTTCTGAATCTATAGCTTCTAACAAAGTAATTAACGGTAGTGCAGTAGAAAAAGTAATTTATGTAATAAATCTAGTCACAGATAGATCTGCTCAGTATTGGTGCGAAGGGCACACAACAAATTTCACATTGCTTTCCACGAACAAAATTGTCGTTAATCCAAGAGGCAATGATGTTCACGTATTTTCTGTTACTGTTGAATACTTAATAATGAATGAAATTGAAGACGAAACTGATCTGAATAGTGTAATTCTGTTACTAACTACTGTATTTAACGCAAACAAAGTACTACTAATGGATATTTTAGAATATAATAAAGATAGCATTTTAATATTATTTCATATTCACGTAGTCCTTGAAGATGCTCACAAAGACTACGGACTAAACATAATAGAAAAAATGAATTTTATAAAAAATCAAGCTAATTTGCATTTGCCCCAACATAATTTAACATTGTTCAACATATCGAGTTCGTTGTATTGTCTACCGACCACCTCTAATGACACCATAATGCTTGATTGGGATTTGACATCAATAGGTCAAATTTCAGCACCAAAACAGTTTTGTTTGCAGAGCAATGGCTTACCAGTTAAGCGGCGCTGTCTTGGATCTTATTTGTATGGAAGCAATTGGGGTAGTATTGATGGGATATGCGATACAGCATACAAACCTTCTACAACAACACAATTTTTATACAACTTTTTAAAGGGACAGGAACCCACTTATACTTCTCGTTTTATCACGGATGGGCTAAATTTTGTTCTGAACGACACAGATATCATAATACCAGCAGATATATACTACCTATCCATGTCACTACGACAAGTTTTACAAATTTCTCATGAACAAGAAATCCTTATCGATATGGGAGACATTGAAAATATAGTGTGGGCGATGGATAGAATGATGACATTGGATTGTGAGTACTTGCGCTTAGCACAAACGCTCAATTCGACTAATGTTATATTAAGTTCGGTAAATTATATTATTGAAAATATTGCGTACAACAATCTGACATCACATCGGAACATTTTAGAAAACGAAAACTATCAAATTGCTGTAAAGCCGAATTTTTTAGTACAAATATCGTTCCCAGTACTGAATAACATAACTGGAATAGCTCTTTCTAAAAACACAGAGTCAGATTCTTTTAATGACATGAAAATTATACCGCTTTATAAAAACACTACACTAGACTACGTGCAATCCATTGAAAACCTGGAAGTCGCTACTTGGGTACCAGAAAACGTGATAAATACTTTGATACCTAATTCTAATGGAAGCGTTCAAAATTATACCAATAAGAAAGACATCAACATTATCCTGAGCTTATTTCATACTGACGCTGTATTTCCGGAAGTAAACTTCAAAGAACATGCCATAAATAGCCGCATTGTAGGTGTCACTATACCAGGATTTATTTCAAACTTAGAATACCCGGTACCCCTTATATTCAGAGAATTCAATGAAAGCAGTTCGAACAAATTTTGTGGATACTGGGATTTTAAGCCACACGGACCTTTCAATAATAATCCTGGGGCGTGGAAAAATGAAGGATGCTACTTAATAAGGTCTGAAGACAAATTAACTGTTTGTGAATGCTATCATCTTACACATTTCGGTCAACTTCTTCATATTTCAGACTACAATTTTCCTAGTAATAATAAAATTCACAAAAGGGCATTAAACATTATCACTTTGATTGGTTGCTTCCTTTCGTTAGGAGGCATAGCCGGTATTTGGATCACGGCCCTAATTTTCGGTAATTGGCGTAAAAAACCTGGTACGAAGGTACTTTTACAGCTGTCGACTTCTATCGCTTTTCCATTGCTTTTCATGGTAGTTTATGATTTGGATAACACGATATTTATTACAGAAAACGATGAACATTTCGTTGACGAAGATTATAGAACTGTTTGTACTGTATTGGGTGCTATACTACATTACTCTATTCTTAGTAGTTTCTTGTGGATGCTAATTGCGGCTGTTCTTCAGTTCGTGCGTTATGTCAGAGTGCTAGGTATACGCAGATCTCCAACGTTCATGATAAAAATAAGTATAGCTGGTTGGGGTGTGCCAATAATACCGGTTGCAATAACACTCATTATCGACCCATCAAATTACATACCTGCTCCGTCCCAAACTAGAGAGATTTGCTATCCTCAAGACTTTGCTTTAGTATTTGGTATTATAATGCCCGTTGGTGTAATACTGATTATAAACGTTATCCTTTTTATACTAGTACTTCACGCTATATCGAGAAACTCTGATGAAGGCACGAAGAATATGGACATGGACCTTGTAGGAGCCCAATTGAGATTATCAGTATTTTTATTTTTCTTGTTGGGTTTGACATGGGTATTTGGCGTCTTATCATTCTCGAAAAATTTACTTTGGTCTTATTTGTTTTGTTTGACTTCTACATTGCAAGGTTTTGTTTTATTTATTTACTTTATTATTTGTGATCCAGTTACTAGAAAAATGTGGGTCGCACTAGTCAAATCACCAGCGAGCCGAGGTAATACTAGAAACAGTATCACAAGTCTAAGCGTGACTTAA
Protein
MNFVFIILILGWTFMTTVQDCPEEISHSGISVKWKQSSWANHIESDPQCYRNQNLITRNCTDNVWKPPLEAVEPCLKVVKNFDVFSCPPGYHKISEKNNDYCYQIGRPSQWEYPCFSSGAASVITDLSELELNSLLQSLVSMNISRYFWLPGKRHKLFNPAVWNIPGPNWGRRVKSNSYLRYRTIFLKNCLLLDVQKRVIITETCSNIYPSLCFYINEFYHPSKCPDGYYALRFQLDNGTCFGIESSDSINGWTFAEFITAKCKNPMNVSSLNALSRFMFKKIAELSKLPNEIWCWLKFTSYYMASKNNTFLEDLEEHNLEGIIDNAGTLGLMNKSRHLSCMACETQVVYLETELMFEYNTVDNNLYLTIYYPSGLWKYNKTDKGVQCFSDANGFVGVINVRELDLFQSKQIISESIASNKVINGSAVEKVIYVINLVTDRSAQYWCEGHTTNFTLLSTNKIVVNPRGNDVHVFSVTVEYLIMNEIEDETDLNSVILLLTTVFNANKVLLMDILEYNKDSILILFHIHVVLEDAHKDYGLNIIEKMNFIKNQANLHLPQHNLTLFNISSSLYCLPTTSNDTIMLDWDLTSIGQISAPKQFCLQSNGLPVKRRCLGSYLYGSNWGSIDGICDTAYKPSTTTQFLYNFLKGQEPTYTSRFITDGLNFVLNDTDIIIPADIYYLSMSLRQVLQISHEQEILIDMGDIENIVWAMDRMMTLDCEYLRLAQTLNSTNVILSSVNYIIENIAYNNLTSHRNILENENYQIAVKPNFLVQISFPVLNNITGIALSKNTESDSFNDMKIIPLYKNTTLDYVQSIENLEVATWVPENVINTLIPNSNGSVQNYTNKKDINIILSLFHTDAVFPEVNFKEHAINSRIVGVTIPGFISNLEYPVPLIFREFNESSSNKFCGYWDFKPHGPFNNNPGAWKNEGCYLIRSEDKLTVCECYHLTHFGQLLHISDYNFPSNNKIHKRALNIITLIGCFLSLGGIAGIWITALIFGNWRKKPGTKVLLQLSTSIAFPLLFMVVYDLDNTIFITENDEHFVDEDYRTVCTVLGAILHYSILSSFLWMLIAAVLQFVRYVRVLGIRRSPTFMIKISIAGWGVPIIPVAITLIIDPSNYIPAPSQTREICYPQDFALVFGIIMPVGVILIINVILFILVLHAISRNSDEGTKNMDMDLVGAQLRLSVFLFFLLGLTWVFGVLSFSKNLLWSYLFCLTSTLQGFVLFIYFIICDPVTRKMWVALVKSPASRGNTRNSITSLSVT
Summary
Similarity
Belongs to the G-protein coupled receptor 2 family.
Uniprot
A0A0L7LMY0
A0A2H1W5L7
A0A1J1J2L0
A0A2P8ZCQ3
A0A336M723
A0A336N2T8
+ More
A0A1B0CWX0 A0A1I8NYF9 A0A1B0CK17 A0A1B0CK19 A0A0L0CGN4 A0A0Q9W7X4 A0A182IYE7 A0A1B0CK20 A0A2C9GPY2 A0A182W434 A0A0Q9WXK3 A0A1I8M7B2 A0A0Q9X474 A0A182Q9Y6 A0A182NAG3 A0A1A9XN02 A0A1B0C2W9 A0A1A9UJW6 A0A1W4X2N8 D6WCP7 A0A1A9W6P3 A0A1B0A8S8 A0A1W4WJ98 A0A1W4WJB1 A0A1Y1NBZ9
A0A1B0CWX0 A0A1I8NYF9 A0A1B0CK17 A0A1B0CK19 A0A0L0CGN4 A0A0Q9W7X4 A0A182IYE7 A0A1B0CK20 A0A2C9GPY2 A0A182W434 A0A0Q9WXK3 A0A1I8M7B2 A0A0Q9X474 A0A182Q9Y6 A0A182NAG3 A0A1A9XN02 A0A1B0C2W9 A0A1A9UJW6 A0A1W4X2N8 D6WCP7 A0A1A9W6P3 A0A1B0A8S8 A0A1W4WJ98 A0A1W4WJB1 A0A1Y1NBZ9
EMBL
JTDY01000526
KOB76802.1
ODYU01006476
SOQ48348.1
CVRI01000066
CRL06028.1
+ More
PYGN01000103 PSN54271.1 UFQS01000650 UFQT01000650 SSX05755.1 SSX26114.1 UFQS01002120 UFQT01002120 SSX13271.1 SSX32708.1 AJWK01032949 AJWK01032950 AJWK01032951 AJWK01015626 AJWK01015627 AJWK01015628 AJWK01015629 AJWK01015631 JRES01000426 KNC31390.1 CH940657 KRF80936.1 AXCP01007276 AJWK01015632 APCN01003972 CH933806 KRG00715.1 KRG00714.1 AXCN02002022 JXJN01024714 KQ971317 EEZ99318.2 GEZM01007006 JAV95482.1
PYGN01000103 PSN54271.1 UFQS01000650 UFQT01000650 SSX05755.1 SSX26114.1 UFQS01002120 UFQT01002120 SSX13271.1 SSX32708.1 AJWK01032949 AJWK01032950 AJWK01032951 AJWK01015626 AJWK01015627 AJWK01015628 AJWK01015629 AJWK01015631 JRES01000426 KNC31390.1 CH940657 KRF80936.1 AXCP01007276 AJWK01015632 APCN01003972 CH933806 KRG00715.1 KRG00714.1 AXCN02002022 JXJN01024714 KQ971317 EEZ99318.2 GEZM01007006 JAV95482.1
Proteomes
Interpro
SUPFAM
SSF56436
SSF56436
Gene 3D
ProteinModelPortal
A0A0L7LMY0
A0A2H1W5L7
A0A1J1J2L0
A0A2P8ZCQ3
A0A336M723
A0A336N2T8
+ More
A0A1B0CWX0 A0A1I8NYF9 A0A1B0CK17 A0A1B0CK19 A0A0L0CGN4 A0A0Q9W7X4 A0A182IYE7 A0A1B0CK20 A0A2C9GPY2 A0A182W434 A0A0Q9WXK3 A0A1I8M7B2 A0A0Q9X474 A0A182Q9Y6 A0A182NAG3 A0A1A9XN02 A0A1B0C2W9 A0A1A9UJW6 A0A1W4X2N8 D6WCP7 A0A1A9W6P3 A0A1B0A8S8 A0A1W4WJ98 A0A1W4WJB1 A0A1Y1NBZ9
A0A1B0CWX0 A0A1I8NYF9 A0A1B0CK17 A0A1B0CK19 A0A0L0CGN4 A0A0Q9W7X4 A0A182IYE7 A0A1B0CK20 A0A2C9GPY2 A0A182W434 A0A0Q9WXK3 A0A1I8M7B2 A0A0Q9X474 A0A182Q9Y6 A0A182NAG3 A0A1A9XN02 A0A1B0C2W9 A0A1A9UJW6 A0A1W4X2N8 D6WCP7 A0A1A9W6P3 A0A1B0A8S8 A0A1W4WJ98 A0A1W4WJB1 A0A1Y1NBZ9
PDB
4DLQ
E-value=0.00721682,
Score=98
Ontologies
GO
Topology
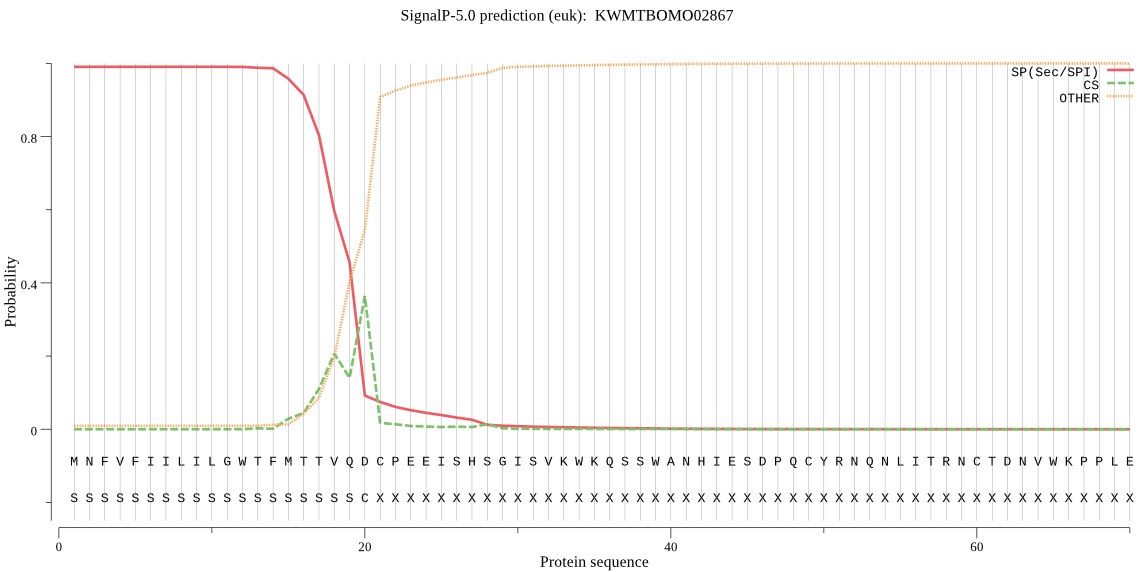
SignalP
Position: 1 - 20,
Likelihood: 0.990475
Length:
1264
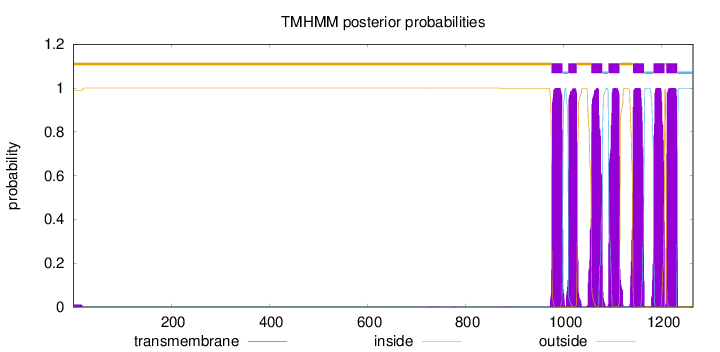
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
155.26998
Exp number, first 60 AAs:
0.19966
Total prob of N-in:
0.01123
outside
1 - 975
TMhelix
976 - 998
inside
999 - 1009
TMhelix
1010 - 1027
outside
1028 - 1056
TMhelix
1057 - 1079
inside
1080 - 1091
TMhelix
1092 - 1114
outside
1115 - 1141
TMhelix
1142 - 1164
inside
1165 - 1183
TMhelix
1184 - 1206
outside
1207 - 1209
TMhelix
1210 - 1232
inside
1233 - 1264
Population Genetic Test Statistics
Pi
152.761624
Theta
170.140916
Tajima's D
-0.766017
CLR
0.524125
CSRT
0.185990700464977
Interpretation
Uncertain
