Gene
KWMTBOMO02864
Pre Gene Modal
BGIBMGA003662
Annotation
PREDICTED:_T-complex-associated_testis-expressed_protein_1-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.036
Sequence
CDS
ATGAAGATTCCATATCTTGTATCCCAGAATACAAGGGAAGCTTACGCTACATCGCATGAAATGAAGACTTCTATTGCTGAAGCCAAGCGTAAAATTGTGGCTGAAGATTTCGAATGGGATGATGGCCTTACTCCAGCACCCCTTAGCTACTTTGCCGCTCTTAGTTTGTCGAGAGCATTTCATATTATTAATCCATTAAACAAAATACTTCAGAAAGATGTTGATATTCTAATGGATTTGTATCCGGCTGATATTCCCTTGAAATGTAGTGTGCCGCTAATACAGGATGAACAATACTGGAAAAGATCTTATGCCACTAAGTTTCCGTCCAGAAAGGAGTTGAACGTGCCATTTTGGAAAGGGAAATTCTTGTCAGCATGTCTACAAGACTATTTAGAAAACGTTAACCCTGATGTCTTTATAGAAGAAGATGCTGTAGAATTGGCAACTTTGGTTAGTCCCTACATTTATGAATTGGGGTTAAATCAACTACGCATGGTTCGGCAACCGACCCCACCAGTCCCTTTTGGGAATATTGTTGAAGACACGTGCCCTTATAGCGTCCCTAAATTATTCGATCACGTGCCTCTGGAACCGGTATTGGTTAAACTTTACAACCTCCAAGAGATATCTTTAGTTTTTGGACTCAATAATATTGAAGACGATTATTTACCAAGATACTTTGAATTTTCAATGGGAGACTGCGAATCTTTATGTCGTGGCCTAGAAGATTTACGACACCTCAAATTGTTTCGGATCAACCGAAGCAATCTAGACTGCTCGAAAGTAAAATTGATTCTACAGAATCTTGTAAAAAAAGAAAGCATTGAGGAGCTTGACTTCAACCATTGTAAGATCGGCGATTATGGAATGAAAGCTCTCGGAGGTTTCATTTACATTCACAAAAATTTAAAAAGAGTGAGAATTGCTAATAACCGATTTAAAGAGGTTGGCGTTCAGGGAATAGCTTATGCCTTGCAAATGAAACCTGCGGCTCCAATAGAACTTTTAGATTTTAGCCTGAATCCAATGTCTTTGGAATGTGCAACAATCTTTGCCGCTGCGTTCGTACGCTGCAAGGAAAAGCCACAAACGTTAATCATCAACTCTTGTGGCTTCAAAGGAGAATCTGCTGAGAAGATTGCAGGATTAATAAGCTTAAATCGAGGTCTGACCAGATTAGATTTGGCAGCTAATGACCTATCGGGAGAAGCTGAAAGAACTTTGGTCAAAGCCGTCGAACAGAACAGAAATATTCTTAGAATAGATTTAAGAAGGACACATATTTCAGAAGAAACTCAATGCAAGATCGACAAACTGTTGGAAAGAAACAAGAACTTAATTGGTCAAGACAGGGAACCGAGCGACGAGATACCGCCTCTTTACCTAAACGAACCTGAATATTTAATCTGGGAATACAATCCCAATAACCCCGACTACATACCCGGCCGATACCCGGAACGAGTTCCAGAAGTTTAA
Protein
MKIPYLVSQNTREAYATSHEMKTSIAEAKRKIVAEDFEWDDGLTPAPLSYFAALSLSRAFHIINPLNKILQKDVDILMDLYPADIPLKCSVPLIQDEQYWKRSYATKFPSRKELNVPFWKGKFLSACLQDYLENVNPDVFIEEDAVELATLVSPYIYELGLNQLRMVRQPTPPVPFGNIVEDTCPYSVPKLFDHVPLEPVLVKLYNLQEISLVFGLNNIEDDYLPRYFEFSMGDCESLCRGLEDLRHLKLFRINRSNLDCSKVKLILQNLVKKESIEELDFNHCKIGDYGMKALGGFIYIHKNLKRVRIANNRFKEVGVQGIAYALQMKPAAPIELLDFSLNPMSLECATIFAAAFVRCKEKPQTLIINSCGFKGESAEKIAGLISLNRGLTRLDLAANDLSGEAERTLVKAVEQNRNILRIDLRRTHISEETQCKIDKLLERNKNLIGQDREPSDEIPPLYLNEPEYLIWEYNPNNPDYIPGRYPERVPEV
Summary
Uniprot
H9J2C5
A0A2W1BTS7
A0A212EX19
A0A2A4K2H5
A0A0L7LMM0
A0A2H1VHP9
+ More
A0A194QTQ6 A0A194PV26 A0A2J7R5R2 A0A1W4X4J4 A0A139WMJ6 N6T3I0 A0A195EVN6 A0A067R9S9 A0A158NAV5 A0A151I165 A0A1Y1LUL5 A0A151X5H6 A0A0L7RCE7 A0A0J7L1F5 A0A195CK12 A0A310SCG2 A0A0C9RTX1 A0A3L8DF33 A0A0N0BCH0 F4X233 A0A1B6GD24 E0VYH5 A0A154PT31 A0A026WH02 A0A2H1WIQ8 A0A2P8YY04 A0A0L7L8G1 A0A2A3EJA9 A0A087ZQY5 A0A369RYL2 B3S841 A0A1I8IEW6 W5LR08 A0A1S8X579 A0A267G5C6 A0A267E315 A0A074ZCS7 G3HC80 A0A3Q0CNM2 D6WNC5 A0A2C9K3T4 A0A2A4JB75 R7TLH4 S9XYF9 A0A1S3AMR9 A0A2D0Q2I2 A0A3B1J346 H2KRI1 A0A1W4Z648 A0A0P7TL15 G3TCX4 A0A2D0Q3F4 I3ML42 A0A3N0YAA3 E2AAL1 A0A212F4C9 K1R202 A0A2Y9DRG7 W4XTU2 A0A1A6FWY2 A0A337SD71 D2GZA2 A0A210R5Q9 F1RQT8 H0XSC5 W5PIM9 A0A2T7NWW4 L8ICG5 H0V1X1 A0A2K6FXA1 A0A2Y9ERF1 W5NHZ4 A0A212D5U7 A0A1U7UM03 F6ZXM2 A0A2Y9P6D7 I3IS38 F1QTI2 A0A3P8Y8S7 M3YMB7 A0A2U3WHJ4 D3ZQW5 A0A340XE42 A0A384B3I8 A0A151P2V9 A0A2Y9JMU2 A0A3P4LTT8 A0A2C9GR26 A0A183AEF1 G1P4D3 A0A2G8LB93 A0A3B1J5Y8
A0A194QTQ6 A0A194PV26 A0A2J7R5R2 A0A1W4X4J4 A0A139WMJ6 N6T3I0 A0A195EVN6 A0A067R9S9 A0A158NAV5 A0A151I165 A0A1Y1LUL5 A0A151X5H6 A0A0L7RCE7 A0A0J7L1F5 A0A195CK12 A0A310SCG2 A0A0C9RTX1 A0A3L8DF33 A0A0N0BCH0 F4X233 A0A1B6GD24 E0VYH5 A0A154PT31 A0A026WH02 A0A2H1WIQ8 A0A2P8YY04 A0A0L7L8G1 A0A2A3EJA9 A0A087ZQY5 A0A369RYL2 B3S841 A0A1I8IEW6 W5LR08 A0A1S8X579 A0A267G5C6 A0A267E315 A0A074ZCS7 G3HC80 A0A3Q0CNM2 D6WNC5 A0A2C9K3T4 A0A2A4JB75 R7TLH4 S9XYF9 A0A1S3AMR9 A0A2D0Q2I2 A0A3B1J346 H2KRI1 A0A1W4Z648 A0A0P7TL15 G3TCX4 A0A2D0Q3F4 I3ML42 A0A3N0YAA3 E2AAL1 A0A212F4C9 K1R202 A0A2Y9DRG7 W4XTU2 A0A1A6FWY2 A0A337SD71 D2GZA2 A0A210R5Q9 F1RQT8 H0XSC5 W5PIM9 A0A2T7NWW4 L8ICG5 H0V1X1 A0A2K6FXA1 A0A2Y9ERF1 W5NHZ4 A0A212D5U7 A0A1U7UM03 F6ZXM2 A0A2Y9P6D7 I3IS38 F1QTI2 A0A3P8Y8S7 M3YMB7 A0A2U3WHJ4 D3ZQW5 A0A340XE42 A0A384B3I8 A0A151P2V9 A0A2Y9JMU2 A0A3P4LTT8 A0A2C9GR26 A0A183AEF1 G1P4D3 A0A2G8LB93 A0A3B1J5Y8
Pubmed
19121390
28756777
22118469
26227816
26354079
18362917
+ More
19820115 23537049 24845553 21347285 28004739 30249741 21719571 20566863 24508170 29403074 30042472 18719581 26392545 25329095 21804562 29704459 28071753 15562597 23254933 23149746 22023798 20798317 22992520 17975172 20010809 28812685 30723633 20809919 22751099 21993624 18464734 23594743 25069045 15057822 15632090 22673903 22293439 29023486
19820115 23537049 24845553 21347285 28004739 30249741 21719571 20566863 24508170 29403074 30042472 18719581 26392545 25329095 21804562 29704459 28071753 15562597 23254933 23149746 22023798 20798317 22992520 17975172 20010809 28812685 30723633 20809919 22751099 21993624 18464734 23594743 25069045 15057822 15632090 22673903 22293439 29023486
EMBL
BABH01007703
KZ149903
PZC78419.1
AGBW02011843
OWR46046.1
NWSH01000238
+ More
PCG78104.1 JTDY01000526 KOB76803.1 ODYU01002619 SOQ40355.1 KQ461150 KPJ08694.1 KQ459597 KPI94985.1 NEVH01006990 PNF36174.1 KQ971312 KYB29096.1 APGK01053935 KB741239 KB632275 ENN72128.1 ERL91102.1 KQ981954 KYN32206.1 KK852832 KDR15318.1 ADTU01010303 ADTU01010304 ADTU01010305 ADTU01010306 KQ976600 KYM79451.1 GEZM01047851 JAV76721.1 KQ982519 KYQ55518.1 KQ414616 KOC68411.1 LBMM01001441 KMQ96319.1 KQ977642 KYN01070.1 KQ769179 OAD52963.1 GBYB01011031 JAG80798.1 QOIP01000009 RLU19080.1 KQ435911 KOX68938.1 GL888565 EGI59449.1 GECZ01009458 JAS60311.1 DS235845 EEB18430.1 KQ435207 KZC15071.1 KK107231 EZA54956.1 ODYU01008934 SOQ52955.1 PYGN01000293 PSN49122.1 JTDY01002365 KOB71571.1 KZ288226 PBC31865.1 NOWV01000128 RDD39630.1 DS985255 EDV21037.1 KV891983 OON21816.1 NIVC01000576 PAA80519.1 NIVC01002775 PAA55297.1 KL596850 KER23432.1 JH000276 RAZU01000009 EGW01340.1 RLQ79504.1 KQ971343 EFA04350.1 NWSH01002160 PCG69016.1 AMQN01012230 AMQN01012231 AMQN01012232 AMQN01012233 KB309387 ELT94524.1 KB017589 EPY76715.1 DF143171 GAA29628.2 JARO02009484 KPP61583.1 AGTP01010790 AGTP01010791 RJVU01048958 ROL43034.1 GL438129 EFN69518.1 AGBW02010386 OWR48587.1 JH818024 EKC39838.1 AAGJ04057178 LZPO01116605 OBS58413.1 AANG04003966 ACTA01184071 GL192391 EFB21282.1 NEDP02000201 OWF56407.1 AEMK02000055 DQIR01029261 HCZ84736.1 AAQR03061276 AMGL01058706 PZQS01000008 PVD25658.1 JH881611 ELR53224.1 AAKN02021390 AHAT01008937 MKHE01000007 OWK13494.1 CU660013 AEYP01006062 AC096454 CH473987 EDM18734.1 AKHW03001146 KYO43408.1 CYRY02001614 VCW66241.1 APCN01000241 UZAN01042188 VDP75249.1 AAPE02036885 AAPE02036886 MRZV01000140 PIK57504.1
PCG78104.1 JTDY01000526 KOB76803.1 ODYU01002619 SOQ40355.1 KQ461150 KPJ08694.1 KQ459597 KPI94985.1 NEVH01006990 PNF36174.1 KQ971312 KYB29096.1 APGK01053935 KB741239 KB632275 ENN72128.1 ERL91102.1 KQ981954 KYN32206.1 KK852832 KDR15318.1 ADTU01010303 ADTU01010304 ADTU01010305 ADTU01010306 KQ976600 KYM79451.1 GEZM01047851 JAV76721.1 KQ982519 KYQ55518.1 KQ414616 KOC68411.1 LBMM01001441 KMQ96319.1 KQ977642 KYN01070.1 KQ769179 OAD52963.1 GBYB01011031 JAG80798.1 QOIP01000009 RLU19080.1 KQ435911 KOX68938.1 GL888565 EGI59449.1 GECZ01009458 JAS60311.1 DS235845 EEB18430.1 KQ435207 KZC15071.1 KK107231 EZA54956.1 ODYU01008934 SOQ52955.1 PYGN01000293 PSN49122.1 JTDY01002365 KOB71571.1 KZ288226 PBC31865.1 NOWV01000128 RDD39630.1 DS985255 EDV21037.1 KV891983 OON21816.1 NIVC01000576 PAA80519.1 NIVC01002775 PAA55297.1 KL596850 KER23432.1 JH000276 RAZU01000009 EGW01340.1 RLQ79504.1 KQ971343 EFA04350.1 NWSH01002160 PCG69016.1 AMQN01012230 AMQN01012231 AMQN01012232 AMQN01012233 KB309387 ELT94524.1 KB017589 EPY76715.1 DF143171 GAA29628.2 JARO02009484 KPP61583.1 AGTP01010790 AGTP01010791 RJVU01048958 ROL43034.1 GL438129 EFN69518.1 AGBW02010386 OWR48587.1 JH818024 EKC39838.1 AAGJ04057178 LZPO01116605 OBS58413.1 AANG04003966 ACTA01184071 GL192391 EFB21282.1 NEDP02000201 OWF56407.1 AEMK02000055 DQIR01029261 HCZ84736.1 AAQR03061276 AMGL01058706 PZQS01000008 PVD25658.1 JH881611 ELR53224.1 AAKN02021390 AHAT01008937 MKHE01000007 OWK13494.1 CU660013 AEYP01006062 AC096454 CH473987 EDM18734.1 AKHW03001146 KYO43408.1 CYRY02001614 VCW66241.1 APCN01000241 UZAN01042188 VDP75249.1 AAPE02036885 AAPE02036886 MRZV01000140 PIK57504.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000037510
UP000053240
UP000053268
+ More
UP000235965 UP000192223 UP000007266 UP000019118 UP000030742 UP000078541 UP000027135 UP000005205 UP000078540 UP000075809 UP000053825 UP000036403 UP000078542 UP000279307 UP000053105 UP000007755 UP000009046 UP000076502 UP000053097 UP000245037 UP000242457 UP000005203 UP000253843 UP000009022 UP000095280 UP000018467 UP000215902 UP000001075 UP000273346 UP000189706 UP000076420 UP000014760 UP000079721 UP000221080 UP000192224 UP000034805 UP000007646 UP000005215 UP000000311 UP000005408 UP000248480 UP000007110 UP000092124 UP000011712 UP000008912 UP000242188 UP000008227 UP000005225 UP000002356 UP000245119 UP000005447 UP000233160 UP000248484 UP000018468 UP000189704 UP000002279 UP000248483 UP000000437 UP000265140 UP000000715 UP000245340 UP000002494 UP000265300 UP000261681 UP000050525 UP000248482 UP000075840 UP000050740 UP000001074 UP000230750
UP000235965 UP000192223 UP000007266 UP000019118 UP000030742 UP000078541 UP000027135 UP000005205 UP000078540 UP000075809 UP000053825 UP000036403 UP000078542 UP000279307 UP000053105 UP000007755 UP000009046 UP000076502 UP000053097 UP000245037 UP000242457 UP000005203 UP000253843 UP000009022 UP000095280 UP000018467 UP000215902 UP000001075 UP000273346 UP000189706 UP000076420 UP000014760 UP000079721 UP000221080 UP000192224 UP000034805 UP000007646 UP000005215 UP000000311 UP000005408 UP000248480 UP000007110 UP000092124 UP000011712 UP000008912 UP000242188 UP000008227 UP000005225 UP000002356 UP000245119 UP000005447 UP000233160 UP000248484 UP000018468 UP000189704 UP000002279 UP000248483 UP000000437 UP000265140 UP000000715 UP000245340 UP000002494 UP000265300 UP000261681 UP000050525 UP000248482 UP000075840 UP000050740 UP000001074 UP000230750
Interpro
Gene 3D
ProteinModelPortal
H9J2C5
A0A2W1BTS7
A0A212EX19
A0A2A4K2H5
A0A0L7LMM0
A0A2H1VHP9
+ More
A0A194QTQ6 A0A194PV26 A0A2J7R5R2 A0A1W4X4J4 A0A139WMJ6 N6T3I0 A0A195EVN6 A0A067R9S9 A0A158NAV5 A0A151I165 A0A1Y1LUL5 A0A151X5H6 A0A0L7RCE7 A0A0J7L1F5 A0A195CK12 A0A310SCG2 A0A0C9RTX1 A0A3L8DF33 A0A0N0BCH0 F4X233 A0A1B6GD24 E0VYH5 A0A154PT31 A0A026WH02 A0A2H1WIQ8 A0A2P8YY04 A0A0L7L8G1 A0A2A3EJA9 A0A087ZQY5 A0A369RYL2 B3S841 A0A1I8IEW6 W5LR08 A0A1S8X579 A0A267G5C6 A0A267E315 A0A074ZCS7 G3HC80 A0A3Q0CNM2 D6WNC5 A0A2C9K3T4 A0A2A4JB75 R7TLH4 S9XYF9 A0A1S3AMR9 A0A2D0Q2I2 A0A3B1J346 H2KRI1 A0A1W4Z648 A0A0P7TL15 G3TCX4 A0A2D0Q3F4 I3ML42 A0A3N0YAA3 E2AAL1 A0A212F4C9 K1R202 A0A2Y9DRG7 W4XTU2 A0A1A6FWY2 A0A337SD71 D2GZA2 A0A210R5Q9 F1RQT8 H0XSC5 W5PIM9 A0A2T7NWW4 L8ICG5 H0V1X1 A0A2K6FXA1 A0A2Y9ERF1 W5NHZ4 A0A212D5U7 A0A1U7UM03 F6ZXM2 A0A2Y9P6D7 I3IS38 F1QTI2 A0A3P8Y8S7 M3YMB7 A0A2U3WHJ4 D3ZQW5 A0A340XE42 A0A384B3I8 A0A151P2V9 A0A2Y9JMU2 A0A3P4LTT8 A0A2C9GR26 A0A183AEF1 G1P4D3 A0A2G8LB93 A0A3B1J5Y8
A0A194QTQ6 A0A194PV26 A0A2J7R5R2 A0A1W4X4J4 A0A139WMJ6 N6T3I0 A0A195EVN6 A0A067R9S9 A0A158NAV5 A0A151I165 A0A1Y1LUL5 A0A151X5H6 A0A0L7RCE7 A0A0J7L1F5 A0A195CK12 A0A310SCG2 A0A0C9RTX1 A0A3L8DF33 A0A0N0BCH0 F4X233 A0A1B6GD24 E0VYH5 A0A154PT31 A0A026WH02 A0A2H1WIQ8 A0A2P8YY04 A0A0L7L8G1 A0A2A3EJA9 A0A087ZQY5 A0A369RYL2 B3S841 A0A1I8IEW6 W5LR08 A0A1S8X579 A0A267G5C6 A0A267E315 A0A074ZCS7 G3HC80 A0A3Q0CNM2 D6WNC5 A0A2C9K3T4 A0A2A4JB75 R7TLH4 S9XYF9 A0A1S3AMR9 A0A2D0Q2I2 A0A3B1J346 H2KRI1 A0A1W4Z648 A0A0P7TL15 G3TCX4 A0A2D0Q3F4 I3ML42 A0A3N0YAA3 E2AAL1 A0A212F4C9 K1R202 A0A2Y9DRG7 W4XTU2 A0A1A6FWY2 A0A337SD71 D2GZA2 A0A210R5Q9 F1RQT8 H0XSC5 W5PIM9 A0A2T7NWW4 L8ICG5 H0V1X1 A0A2K6FXA1 A0A2Y9ERF1 W5NHZ4 A0A212D5U7 A0A1U7UM03 F6ZXM2 A0A2Y9P6D7 I3IS38 F1QTI2 A0A3P8Y8S7 M3YMB7 A0A2U3WHJ4 D3ZQW5 A0A340XE42 A0A384B3I8 A0A151P2V9 A0A2Y9JMU2 A0A3P4LTT8 A0A2C9GR26 A0A183AEF1 G1P4D3 A0A2G8LB93 A0A3B1J5Y8
PDB
4PEQ
E-value=2.04439e-08,
Score=142
Ontologies
GO
Topology
Subcellular location
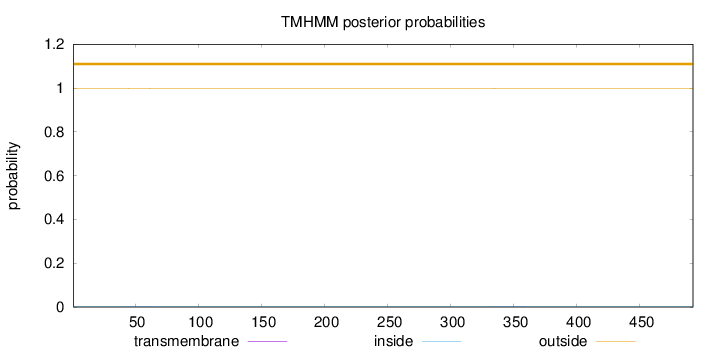
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0231
Exp number, first 60 AAs:
0.00462
Total prob of N-in:
0.00260
outside
1 - 492
Population Genetic Test Statistics
Pi
138.056252
Theta
128.230829
Tajima's D
-0.462005
CLR
0.740692
CSRT
0.251737413129344
Interpretation
Possibly Positive selection
