Pre Gene Modal
BGIBMGA006777
Annotation
cdc2-related_kinase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.975
Sequence
CDS
ATGTCTCAAGGTTCGGAAAAATCTGCATCTAATCATGACGTGGTGGATCCTACAGGTCCTGGGACGAGAAAAGGTGTTCTTATATCTTTCAGAACGGGAAAAACAATGGAAATACCAGAAAAAGATATATTAGGAAAATGTCGTTTTGTGGGAGAATTCGAAAAATTAAATCGCATTGGTGAAGGCACATACGGAATAGTCTACCGGGCTAAGAACAAGGCAAATGGCAGCATAGTTGCTTTGAAAAAGGTCAGAATGGATGTGGAGAAGGATGGTCTACCGCTGAGTGGCCTTCGTGAGATTCAAGTGCTAATGTCATGTCGACATGAGAACATTGTGCAACTCAAAGAGGTGTTAGTTGGTCGTTCACTTGAGAGCATTTTTCTCTCAATGGAGTACTGCGAACAAGATTTAGCCTCATTACTTGATAATATGTCTTCACCTTTTACGGAATCTCAAGTCAAATGTTTGATGCTGCAAGTATTGAAGGGTCTAAAGTATTTGCATTCCAACTTCATTGTGCATAGGGACTTGAAGGTGTCCAATCTTCTTTTAACTGATAAAGGCTGTGTCAAAATAGCTGATTTCGGTCTAGCTCGTTGGTTAGGCGCCCCGGCCAGAAGTGCAACGCCTCGTGTTGTCACCTTATGGTATAGGGCGCCCGAATTACTTCTGCAGTCACCACGACAGACACCTGCACTGGATATGTGGGCAGCTGGATGCATTCTTGGAGAACTTCTTGCCAACAAACCATTGCTCCCCGGAAGAACCGAGATAGAACAGCTTGAACTTATCGTGGATTTACTTGGCACCCCATCGGATGCCATTTGGCCAGAGTTTAGTGCACTGCCAGCGTTGCAGAACTTCACTCTGAAACAGCAGCCATACAATAATTTGAAGCAGAGGTTCCCGTGGCTCTCGGCTGCCGGCTTACGGTTACTGAACTTCCTATTCATGTACGATCCCAATAAACGTGCCACGGCCGAAGAGTGCCTACAGAGTTCATACTTTAAAGAAGAACCTTTACCGTGCGATCCGAAATTAATGCCCAGCTTCCCACAACATAGAAATATGAAAGGACAAACAAAGGCGACGTCAAATCAATTGAATATACCACTGTCCAATCTTAACACGGGAGCGAACGACCAAACAAACAATCTACCAGCTATTTCAGATTTACTGGGGTCGCTGGTCAAGAAACGCAGACTAGAGTAG
Protein
MSQGSEKSASNHDVVDPTGPGTRKGVLISFRTGKTMEIPEKDILGKCRFVGEFEKLNRIGEGTYGIVYRAKNKANGSIVALKKVRMDVEKDGLPLSGLREIQVLMSCRHENIVQLKEVLVGRSLESIFLSMEYCEQDLASLLDNMSSPFTESQVKCLMLQVLKGLKYLHSNFIVHRDLKVSNLLLTDKGCVKIADFGLARWLGAPARSATPRVVTLWYRAPELLLQSPRQTPALDMWAAGCILGELLANKPLLPGRTEIEQLELIVDLLGTPSDAIWPEFSALPALQNFTLKQQPYNNLKQRFPWLSAAGLRLLNFLFMYDPNKRATAEECLQSSYFKEEPLPCDPKLMPSFPQHRNMKGQTKATSNQLNIPLSNLNTGANDQTNNLPAISDLLGSLVKKRRLE
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
O17508
H9JB82
A0A2W1BXN9
A0A2A4J502
V5K6L7
A0A2H1W6U9
+ More
S4NY05 A0A212F1C9 I4DL22 A0A194PNU1 A0A1Y1KB46 A0A1W4X545 A0A232ELX8 K7J422 D6WH04 A0A158NSQ2 A0A1W4XFG2 A0A026WWN6 A0A2A3EIH3 A0A088ATD3 A0A310SDI6 A0A291S6V2 N6T0B9 A0A0L7RK38 A0A2J7QCS8 A0A2P8Y3V1 F4W888 A0A151HYR3 A0A195CAV8 A0A195EXL3 E2AI11 R4WSN6 A0A224XPI9 R4G3D3 A0A069DTX7 U4UC43 A0A1B6CMG3 A0A0A9YBM0 E0VQN1 A0A0V0G947 A0A154NXA4 A0A023F8Z3 V9ILL2 A0A0A9XHK3 A0A195DUR1 A0A151X041 A0A067QKM2 A0A0P4VPI9 A0A1B6EK72 A0A0L0BSA1 D3TRH8 A0A1A9VQW0 A0A1A9Z0L7 A0A1I8NAN6 A0A1A9W0T8 A0A1I8Q2A7 A0A1Q3F540 O61440 U5ETN2 A0A182H3B1 B0W3P6 A0A1B0GBI5 A0A0P6IXT8 A0A034V192 A0A0N8C7I2 A0A0K8W8K4 A0A0P5FZL4 A0A1S4F9X3 A0A0A1WGN3 Q17AT5 A0A0K8UGI4 A0A0P6AJ80 A0A0P5R817 E9G4D7 A0A3L8D954 A0A182ITQ7 W5J5I5 A0A1B6JX62 J9JUH5 A0A0P6HEF4 A0A1B0DIR3 A0A0P6IGP1 A0A023GJ98 A0A1B6L2X9 A0A131XLW8 A0A023FLY4 L7M7V6 A0A0C9QEU4 A0A0K8RFK9 A0A2S2PJS9 A0A023FW10 A0A1E1X914 A0A131YFZ7 A0A2R5LK63 A0A131YPL1 A0A224YMI5 A0A224Z6X8 E2C8E3 A0A1Z5LF97 B7P2I5
S4NY05 A0A212F1C9 I4DL22 A0A194PNU1 A0A1Y1KB46 A0A1W4X545 A0A232ELX8 K7J422 D6WH04 A0A158NSQ2 A0A1W4XFG2 A0A026WWN6 A0A2A3EIH3 A0A088ATD3 A0A310SDI6 A0A291S6V2 N6T0B9 A0A0L7RK38 A0A2J7QCS8 A0A2P8Y3V1 F4W888 A0A151HYR3 A0A195CAV8 A0A195EXL3 E2AI11 R4WSN6 A0A224XPI9 R4G3D3 A0A069DTX7 U4UC43 A0A1B6CMG3 A0A0A9YBM0 E0VQN1 A0A0V0G947 A0A154NXA4 A0A023F8Z3 V9ILL2 A0A0A9XHK3 A0A195DUR1 A0A151X041 A0A067QKM2 A0A0P4VPI9 A0A1B6EK72 A0A0L0BSA1 D3TRH8 A0A1A9VQW0 A0A1A9Z0L7 A0A1I8NAN6 A0A1A9W0T8 A0A1I8Q2A7 A0A1Q3F540 O61440 U5ETN2 A0A182H3B1 B0W3P6 A0A1B0GBI5 A0A0P6IXT8 A0A034V192 A0A0N8C7I2 A0A0K8W8K4 A0A0P5FZL4 A0A1S4F9X3 A0A0A1WGN3 Q17AT5 A0A0K8UGI4 A0A0P6AJ80 A0A0P5R817 E9G4D7 A0A3L8D954 A0A182ITQ7 W5J5I5 A0A1B6JX62 J9JUH5 A0A0P6HEF4 A0A1B0DIR3 A0A0P6IGP1 A0A023GJ98 A0A1B6L2X9 A0A131XLW8 A0A023FLY4 L7M7V6 A0A0C9QEU4 A0A0K8RFK9 A0A2S2PJS9 A0A023FW10 A0A1E1X914 A0A131YFZ7 A0A2R5LK63 A0A131YPL1 A0A224YMI5 A0A224Z6X8 E2C8E3 A0A1Z5LF97 B7P2I5
Pubmed
9099577
19121390
28756777
24517229
23622113
22118469
+ More
22651552 26354079 28004739 28648823 20075255 18362917 19820115 21347285 24508170 28992199 23537049 29403074 21719571 20798317 23691247 26334808 25401762 20566863 25474469 24845553 27129103 26108605 20353571 25315136 26483478 26999592 25348373 25830018 17510324 21292972 30249741 20920257 23761445 28049606 25576852 28503490 26830274 28797301 28528879
22651552 26354079 28004739 28648823 20075255 18362917 19820115 21347285 24508170 28992199 23537049 29403074 21719571 20798317 23691247 26334808 25401762 20566863 25474469 24845553 27129103 26108605 20353571 25315136 26483478 26999592 25348373 25830018 17510324 21292972 30249741 20920257 23761445 28049606 25576852 28503490 26830274 28797301 28528879
EMBL
D85135
BAA21484.1
BABH01008744
KZ149903
PZC78415.1
NWSH01003044
+ More
PCG67061.1 KC188798 AGT99005.1 ODYU01006692 SOQ48763.1 GAIX01013990 JAA78570.1 AGBW02010901 OWR47536.1 AK401990 BAM18612.1 KQ459597 KPI94982.1 GEZM01087383 JAV58723.1 NNAY01003487 OXU19341.1 AAZX01001011 KQ971321 EFA00613.1 ADTU01025105 ADTU01025106 KK107078 EZA60472.1 KZ288253 PBC30986.1 KQ797156 OAD46977.1 MF432984 ATL75335.1 APGK01057490 KB741280 ENN70953.1 KQ414578 KOC71173.1 NEVH01015880 PNF26391.1 PYGN01000957 PSN38923.1 GL887898 EGI69583.1 KQ976723 KYM76668.1 KQ978023 KYM97999.1 KQ981948 KYN32637.1 GL439621 EFN66919.1 AK417722 BAN20937.1 GFTR01005944 JAW10482.1 ACPB03013281 GAHY01001781 JAA75729.1 GBGD01001663 JAC87226.1 KB632033 ERL88176.1 GEDC01022723 JAS14575.1 GBHO01013127 GBRD01008701 JAG30477.1 JAG57120.1 DS235430 EEB15687.1 GECL01001711 JAP04413.1 KQ434777 KZC04257.1 GBBI01000795 JAC17917.1 JR051729 AEY61547.1 GBHO01024503 JAG19101.1 KQ980322 KYN16586.1 KQ982624 KYQ53693.1 KK853225 KDR09684.1 GDKW01002924 JAI53671.1 GECZ01031445 JAS38324.1 JRES01001442 KNC22886.1 EZ424030 ADD20306.1 GFDL01012457 JAV22588.1 AF034643 AAC79672.4 GANO01002703 JAB57168.1 JXUM01008935 JXUM01107104 KQ565112 KQ560272 KXJ71326.1 KXJ83396.1 DS231833 EDS32193.1 CCAG010014016 GDUN01000132 JAN95787.1 GAKP01022693 JAC36259.1 GDIQ01107203 JAL44523.1 GDHF01004781 JAI47533.1 GDIP01242665 GDIQ01252707 GDIQ01252706 GDIQ01249659 GDIQ01249658 GDIQ01248256 GDIQ01248255 GDIQ01241407 GDIQ01101507 GDIP01055218 GDIQ01035332 LRGB01000763 JAI80736.1 JAK03469.1 KZS16111.1 GBXI01016491 JAC97800.1 CH477330 EAT43373.1 GDHF01026547 JAI25767.1 GDIP01028610 JAM75105.1 GDIQ01109397 JAL42329.1 GL732532 EFX85276.1 QOIP01000011 RLU16864.1 ADMH02002096 ETN59251.1 GECU01004223 JAT03484.1 ABLF02036611 GDIQ01020385 JAN74352.1 AJVK01015587 GDIQ01005349 JAN89388.1 GBBM01001439 JAC33979.1 GEBQ01021920 JAT18057.1 GEFH01002060 JAP66521.1 GBBK01002794 JAC21688.1 GACK01004653 JAA60381.1 GBYB01001824 JAG71591.1 GADI01004539 JAA69269.1 GGMR01017093 MBY29712.1 GBBL01001319 JAC26001.1 GFAC01003433 JAT95755.1 GEDV01011155 JAP77402.1 GGLE01005692 MBY09818.1 GEDV01007354 JAP81203.1 GFPF01007009 MAA18155.1 GFPF01010974 MAA22120.1 GL453666 EFN75778.1 GFJQ02000903 JAW06067.1 ABJB010004628 ABJB010470419 ABJB010576104 DS622547 EEC00807.1
PCG67061.1 KC188798 AGT99005.1 ODYU01006692 SOQ48763.1 GAIX01013990 JAA78570.1 AGBW02010901 OWR47536.1 AK401990 BAM18612.1 KQ459597 KPI94982.1 GEZM01087383 JAV58723.1 NNAY01003487 OXU19341.1 AAZX01001011 KQ971321 EFA00613.1 ADTU01025105 ADTU01025106 KK107078 EZA60472.1 KZ288253 PBC30986.1 KQ797156 OAD46977.1 MF432984 ATL75335.1 APGK01057490 KB741280 ENN70953.1 KQ414578 KOC71173.1 NEVH01015880 PNF26391.1 PYGN01000957 PSN38923.1 GL887898 EGI69583.1 KQ976723 KYM76668.1 KQ978023 KYM97999.1 KQ981948 KYN32637.1 GL439621 EFN66919.1 AK417722 BAN20937.1 GFTR01005944 JAW10482.1 ACPB03013281 GAHY01001781 JAA75729.1 GBGD01001663 JAC87226.1 KB632033 ERL88176.1 GEDC01022723 JAS14575.1 GBHO01013127 GBRD01008701 JAG30477.1 JAG57120.1 DS235430 EEB15687.1 GECL01001711 JAP04413.1 KQ434777 KZC04257.1 GBBI01000795 JAC17917.1 JR051729 AEY61547.1 GBHO01024503 JAG19101.1 KQ980322 KYN16586.1 KQ982624 KYQ53693.1 KK853225 KDR09684.1 GDKW01002924 JAI53671.1 GECZ01031445 JAS38324.1 JRES01001442 KNC22886.1 EZ424030 ADD20306.1 GFDL01012457 JAV22588.1 AF034643 AAC79672.4 GANO01002703 JAB57168.1 JXUM01008935 JXUM01107104 KQ565112 KQ560272 KXJ71326.1 KXJ83396.1 DS231833 EDS32193.1 CCAG010014016 GDUN01000132 JAN95787.1 GAKP01022693 JAC36259.1 GDIQ01107203 JAL44523.1 GDHF01004781 JAI47533.1 GDIP01242665 GDIQ01252707 GDIQ01252706 GDIQ01249659 GDIQ01249658 GDIQ01248256 GDIQ01248255 GDIQ01241407 GDIQ01101507 GDIP01055218 GDIQ01035332 LRGB01000763 JAI80736.1 JAK03469.1 KZS16111.1 GBXI01016491 JAC97800.1 CH477330 EAT43373.1 GDHF01026547 JAI25767.1 GDIP01028610 JAM75105.1 GDIQ01109397 JAL42329.1 GL732532 EFX85276.1 QOIP01000011 RLU16864.1 ADMH02002096 ETN59251.1 GECU01004223 JAT03484.1 ABLF02036611 GDIQ01020385 JAN74352.1 AJVK01015587 GDIQ01005349 JAN89388.1 GBBM01001439 JAC33979.1 GEBQ01021920 JAT18057.1 GEFH01002060 JAP66521.1 GBBK01002794 JAC21688.1 GACK01004653 JAA60381.1 GBYB01001824 JAG71591.1 GADI01004539 JAA69269.1 GGMR01017093 MBY29712.1 GBBL01001319 JAC26001.1 GFAC01003433 JAT95755.1 GEDV01011155 JAP77402.1 GGLE01005692 MBY09818.1 GEDV01007354 JAP81203.1 GFPF01007009 MAA18155.1 GFPF01010974 MAA22120.1 GL453666 EFN75778.1 GFJQ02000903 JAW06067.1 ABJB010004628 ABJB010470419 ABJB010576104 DS622547 EEC00807.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000192223
UP000215335
+ More
UP000002358 UP000007266 UP000005205 UP000053097 UP000242457 UP000005203 UP000019118 UP000053825 UP000235965 UP000245037 UP000007755 UP000078540 UP000078542 UP000078541 UP000000311 UP000015103 UP000030742 UP000009046 UP000076502 UP000078492 UP000075809 UP000027135 UP000037069 UP000078200 UP000092445 UP000095301 UP000091820 UP000095300 UP000069940 UP000249989 UP000002320 UP000092444 UP000076858 UP000008820 UP000000305 UP000279307 UP000075880 UP000000673 UP000007819 UP000092462 UP000008237 UP000001555
UP000002358 UP000007266 UP000005205 UP000053097 UP000242457 UP000005203 UP000019118 UP000053825 UP000235965 UP000245037 UP000007755 UP000078540 UP000078542 UP000078541 UP000000311 UP000015103 UP000030742 UP000009046 UP000076502 UP000078492 UP000075809 UP000027135 UP000037069 UP000078200 UP000092445 UP000095301 UP000091820 UP000095300 UP000069940 UP000249989 UP000002320 UP000092444 UP000076858 UP000008820 UP000000305 UP000279307 UP000075880 UP000000673 UP000007819 UP000092462 UP000008237 UP000001555
Interpro
IPR011009
Kinase-like_dom_sf
+ More
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR009057 Homeobox-like_sf
IPR001005 SANT/Myb
IPR017930 Myb_dom
IPR002156 RNaseH_domain
IPR002999 Tudor
IPR000504 RRM_dom
IPR038069 Pelota/DOM34_N
IPR005140 eRF1_1_Pelota
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035437 SNase_OB-fold_sf
IPR035979 RBD_domain_sf
IPR002893 Znf_MYND
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR009057 Homeobox-like_sf
IPR001005 SANT/Myb
IPR017930 Myb_dom
IPR002156 RNaseH_domain
IPR002999 Tudor
IPR000504 RRM_dom
IPR038069 Pelota/DOM34_N
IPR005140 eRF1_1_Pelota
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035437 SNase_OB-fold_sf
IPR035979 RBD_domain_sf
IPR002893 Znf_MYND
Gene 3D
ProteinModelPortal
O17508
H9JB82
A0A2W1BXN9
A0A2A4J502
V5K6L7
A0A2H1W6U9
+ More
S4NY05 A0A212F1C9 I4DL22 A0A194PNU1 A0A1Y1KB46 A0A1W4X545 A0A232ELX8 K7J422 D6WH04 A0A158NSQ2 A0A1W4XFG2 A0A026WWN6 A0A2A3EIH3 A0A088ATD3 A0A310SDI6 A0A291S6V2 N6T0B9 A0A0L7RK38 A0A2J7QCS8 A0A2P8Y3V1 F4W888 A0A151HYR3 A0A195CAV8 A0A195EXL3 E2AI11 R4WSN6 A0A224XPI9 R4G3D3 A0A069DTX7 U4UC43 A0A1B6CMG3 A0A0A9YBM0 E0VQN1 A0A0V0G947 A0A154NXA4 A0A023F8Z3 V9ILL2 A0A0A9XHK3 A0A195DUR1 A0A151X041 A0A067QKM2 A0A0P4VPI9 A0A1B6EK72 A0A0L0BSA1 D3TRH8 A0A1A9VQW0 A0A1A9Z0L7 A0A1I8NAN6 A0A1A9W0T8 A0A1I8Q2A7 A0A1Q3F540 O61440 U5ETN2 A0A182H3B1 B0W3P6 A0A1B0GBI5 A0A0P6IXT8 A0A034V192 A0A0N8C7I2 A0A0K8W8K4 A0A0P5FZL4 A0A1S4F9X3 A0A0A1WGN3 Q17AT5 A0A0K8UGI4 A0A0P6AJ80 A0A0P5R817 E9G4D7 A0A3L8D954 A0A182ITQ7 W5J5I5 A0A1B6JX62 J9JUH5 A0A0P6HEF4 A0A1B0DIR3 A0A0P6IGP1 A0A023GJ98 A0A1B6L2X9 A0A131XLW8 A0A023FLY4 L7M7V6 A0A0C9QEU4 A0A0K8RFK9 A0A2S2PJS9 A0A023FW10 A0A1E1X914 A0A131YFZ7 A0A2R5LK63 A0A131YPL1 A0A224YMI5 A0A224Z6X8 E2C8E3 A0A1Z5LF97 B7P2I5
S4NY05 A0A212F1C9 I4DL22 A0A194PNU1 A0A1Y1KB46 A0A1W4X545 A0A232ELX8 K7J422 D6WH04 A0A158NSQ2 A0A1W4XFG2 A0A026WWN6 A0A2A3EIH3 A0A088ATD3 A0A310SDI6 A0A291S6V2 N6T0B9 A0A0L7RK38 A0A2J7QCS8 A0A2P8Y3V1 F4W888 A0A151HYR3 A0A195CAV8 A0A195EXL3 E2AI11 R4WSN6 A0A224XPI9 R4G3D3 A0A069DTX7 U4UC43 A0A1B6CMG3 A0A0A9YBM0 E0VQN1 A0A0V0G947 A0A154NXA4 A0A023F8Z3 V9ILL2 A0A0A9XHK3 A0A195DUR1 A0A151X041 A0A067QKM2 A0A0P4VPI9 A0A1B6EK72 A0A0L0BSA1 D3TRH8 A0A1A9VQW0 A0A1A9Z0L7 A0A1I8NAN6 A0A1A9W0T8 A0A1I8Q2A7 A0A1Q3F540 O61440 U5ETN2 A0A182H3B1 B0W3P6 A0A1B0GBI5 A0A0P6IXT8 A0A034V192 A0A0N8C7I2 A0A0K8W8K4 A0A0P5FZL4 A0A1S4F9X3 A0A0A1WGN3 Q17AT5 A0A0K8UGI4 A0A0P6AJ80 A0A0P5R817 E9G4D7 A0A3L8D954 A0A182ITQ7 W5J5I5 A0A1B6JX62 J9JUH5 A0A0P6HEF4 A0A1B0DIR3 A0A0P6IGP1 A0A023GJ98 A0A1B6L2X9 A0A131XLW8 A0A023FLY4 L7M7V6 A0A0C9QEU4 A0A0K8RFK9 A0A2S2PJS9 A0A023FW10 A0A1E1X914 A0A131YFZ7 A0A2R5LK63 A0A131YPL1 A0A224YMI5 A0A224Z6X8 E2C8E3 A0A1Z5LF97 B7P2I5
PDB
5EFQ
E-value=1.68098e-70,
Score=676
Ontologies
GO
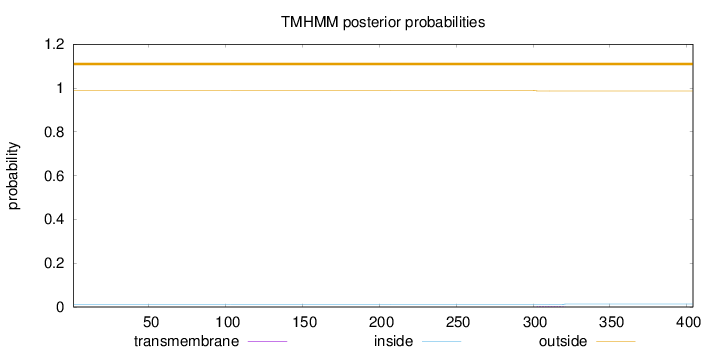
Topology
Length:
404
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04918
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01133
outside
1 - 404
Population Genetic Test Statistics
Pi
166.052404
Theta
147.297133
Tajima's D
0.38436
CLR
0.318795
CSRT
0.487675616219189
Interpretation
Uncertain
