Gene
KWMTBOMO02856
Pre Gene Modal
BGIBMGA003663
Annotation
PREDICTED:_methionine_aminopeptidase_1D?_mitochondrial_isoform_X1_[Bombyx_mori]
Full name
Methionine aminopeptidase
Location in the cell
Cytoplasmic Reliability : 1.432 Mitochondrial Reliability : 1.131
Sequence
CDS
ATGAGGGTGCCGAAAGCCGTTAATCCCAGGAAACGGCTTCAGTTGTTGAAAAAGTTTGGTATTTACGAAAAAGTTTTACCTGTAGAAACAACTCCAAGTCGAGTTGTACCAGATACCATTGAAAGACCTGATTATTTAGCTAGAGGAAAACAATTGACATTACCATGTTTACCGGAGATTAAAGATGATAATCAATTGAAAGGAATGAGAACTAGCTGTAAATTGGCTTCTACTATTCTATCACAAGTGCAAAATTTTATAAAGCCTGGAGTTACAACAGATGACATTGATCAACTAGTCCATTCGTTAATAATTAAAGCTGGAGCATACCCTTCACCACTTCATTATAAGGGATTCCCAAAAAGTATTTGTACATCTGTCAATAATGTGGCTGTGCATGGTATTCCTGATCTAAGGCCACTATCTGATGGGGATATTGTCAATGTTGATATAACAGTATTTTTACATGGTTTTCATGGCGATTGCTCAAAAACTTTCAAAGTTGGTAATGTAGACGACAGAGGTCTACAACTAGTTGAAATCACCGAGCAATGTCTTGGAATCGGAATCGGAACATGCGGGCCAGGAGTACCATTCAGTGAAATCGGCGCCAGCATTCAAAGGCACGCACGTAGAAATGGCCTCACTGTTATACCATCGATTGTGGGTCATGGAATTGGACAGTATTTTCATGGGCCACCAGATATTTATCACATAAGAAACCGTTATCCAGGAGTAATGAAAGCAGGAATGACATTTACAATAGAACCAGTCGTTTCACATGGGACTCAAAATACTGTCTTATTAGATGATGGCTGGACGATTTTAACAGAGGACGGATCGAGAACATCTCAGATCGAACATACCGTATTAATTACTGATGATGGAGTAGAAATCTTAACGAAATGA
Protein
MRVPKAVNPRKRLQLLKKFGIYEKVLPVETTPSRVVPDTIERPDYLARGKQLTLPCLPEIKDDNQLKGMRTSCKLASTILSQVQNFIKPGVTTDDIDQLVHSLIIKAGAYPSPLHYKGFPKSICTSVNNVAVHGIPDLRPLSDGDIVNVDITVFLHGFHGDCSKTFKVGNVDDRGLQLVEITEQCLGIGIGTCGPGVPFSEIGASIQRHARRNGLTVIPSIVGHGIGQYFHGPPDIYHIRNRYPGVMKAGMTFTIEPVVSHGTQNTVLLDDGWTILTEDGSRTSQIEHTVLITDDGVEILTK
Summary
Description
Removes the N-terminal methionine from nascent proteins. The N-terminal methionine is often cleaved when the second residue in the primary sequence is small and uncharged (Met-Ala-, Cys, Gly, Pro, Ser, Thr, or Val).
Catalytic Activity
Release of N-terminal amino acids, preferentially methionine, from peptides and arylamides.
Similarity
Belongs to the peptidase M24A family. Methionine aminopeptidase type 1 subfamily.
Belongs to the peptidase M24A family.
Belongs to the peptidase M24A family.
Feature
chain Methionine aminopeptidase
Uniprot
A0A194QT48
A0A2W1BIT7
A0A2A4J6L5
A0A194PV21
A0A2H1W0H7
A0A139WFT0
+ More
A0A1Y1LLJ2 A0A0T6B2D2 A0A0M9A8W1 A0A2A3EJC5 A0A2J7QKU0 K7IPQ7 A0A158P1G2 A0A310SL33 N6TD21 A0A195F3D6 U4UFB3 A0A232ERQ6 A0A182RLC3 A0A151I2D3 A0A182K2U9 A0A182I7M7 F4WYJ4 A0A195CU66 A0A182QAV1 A0A182NBE8 A0A182SVX6 A0A1B6LTB6 E2BNX3 A0A182VE41 A0A1B6IGF3 A0A182U821 E2AWR2 A0A182LMD7 A0A1B6F8M9 A0A1B6JTD1 E9IJM3 A0A182ISS7 A0A1B6IFU2 Q7PWC9 A0A067QPD7 A0A182GI67 Q16K07 A0A026X1V3 A0A151X7C0 A0A182GBE7 A0A182VU20 B0XHA3 A0A369SEK8 A0A1L8E2W7 A0A0P5P536 A0A0P5J527 B3RPM1 A0A195E2Q4 A0A084VKI8 A0A1W4UT93 A0A1A9W0P3 A0A1Q3FDL9 E0VVC1 A0A1I8NDJ9 A0A1A9UDC1 A0A2J7QKR7 W5J3I3 A0A1I8PHN9 A0A0L0CGW9 A0A1B0FEQ1 D3TL49 A0A1B0DPY8 A0A0V0G995 A0A2S2QR89 A0A0Q9W8A1 B4MUC6 A0A2R7X0H5 A0A0P4WD60 A0A224XV30 A0A226F6J1 A0A1B0BV25 A0A182WXR4 A0A2H8TF84 A0A154P1I2 A0A0Q9W9E6 B4HWP5 A0A0C9QZP1 A0A1W4WW54 A0A2P8XKX7 A0A0P6JSW7 A0A0P6BVH9 A0A0P4VWL1 R4G8V2 A0A0L7RD34 A0A0Q9WFK2 H1UUC3 A0A1W4WKA7 C3XSK2 Q9VKV9 R7UJT7 A0A1B6E9Y8 B4KHR7 A0A069DRZ4
A0A1Y1LLJ2 A0A0T6B2D2 A0A0M9A8W1 A0A2A3EJC5 A0A2J7QKU0 K7IPQ7 A0A158P1G2 A0A310SL33 N6TD21 A0A195F3D6 U4UFB3 A0A232ERQ6 A0A182RLC3 A0A151I2D3 A0A182K2U9 A0A182I7M7 F4WYJ4 A0A195CU66 A0A182QAV1 A0A182NBE8 A0A182SVX6 A0A1B6LTB6 E2BNX3 A0A182VE41 A0A1B6IGF3 A0A182U821 E2AWR2 A0A182LMD7 A0A1B6F8M9 A0A1B6JTD1 E9IJM3 A0A182ISS7 A0A1B6IFU2 Q7PWC9 A0A067QPD7 A0A182GI67 Q16K07 A0A026X1V3 A0A151X7C0 A0A182GBE7 A0A182VU20 B0XHA3 A0A369SEK8 A0A1L8E2W7 A0A0P5P536 A0A0P5J527 B3RPM1 A0A195E2Q4 A0A084VKI8 A0A1W4UT93 A0A1A9W0P3 A0A1Q3FDL9 E0VVC1 A0A1I8NDJ9 A0A1A9UDC1 A0A2J7QKR7 W5J3I3 A0A1I8PHN9 A0A0L0CGW9 A0A1B0FEQ1 D3TL49 A0A1B0DPY8 A0A0V0G995 A0A2S2QR89 A0A0Q9W8A1 B4MUC6 A0A2R7X0H5 A0A0P4WD60 A0A224XV30 A0A226F6J1 A0A1B0BV25 A0A182WXR4 A0A2H8TF84 A0A154P1I2 A0A0Q9W9E6 B4HWP5 A0A0C9QZP1 A0A1W4WW54 A0A2P8XKX7 A0A0P6JSW7 A0A0P6BVH9 A0A0P4VWL1 R4G8V2 A0A0L7RD34 A0A0Q9WFK2 H1UUC3 A0A1W4WKA7 C3XSK2 Q9VKV9 R7UJT7 A0A1B6E9Y8 B4KHR7 A0A069DRZ4
EC Number
3.4.11.18
Pubmed
26354079
28756777
18362917
19820115
28004739
20075255
+ More
21347285 23537049 28648823 21719571 20798317 20966253 21282665 12364791 14747013 17210077 24845553 26483478 17510324 24508170 30249741 30042472 18719581 24438588 20566863 25315136 20920257 23761445 26108605 20353571 17994087 29403074 27129103 18563158 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23254933 26334808
21347285 23537049 28648823 21719571 20798317 20966253 21282665 12364791 14747013 17210077 24845553 26483478 17510324 24508170 30249741 30042472 18719581 24438588 20566863 25315136 20920257 23761445 26108605 20353571 17994087 29403074 27129103 18563158 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23254933 26334808
EMBL
KQ461150
KPJ08698.1
KZ150123
PZC73147.1
NWSH01003044
PCG67063.1
+ More
KQ459597 KPI94980.1 ODYU01005295 SOQ46014.1 KQ971351 KYB26774.1 GEZM01058144 JAV71867.1 LJIG01016105 KRT81599.1 KQ435711 KOX79625.1 KZ288248 PBC31131.1 NEVH01013258 PNF29178.1 ADTU01006535 KQ762882 OAD55381.1 APGK01035263 KB740923 ENN78234.1 KQ981852 KYN34973.1 KB632333 ERL92669.1 NNAY01002575 OXU21017.1 KQ976538 KYM81320.1 APCN01003428 GL888450 EGI60726.1 KQ977305 KYN03679.1 AXCN02000624 GEBQ01013118 JAT26859.1 GL449511 EFN82565.1 GECU01021708 JAS85998.1 GL443403 EFN62138.1 GECZ01023181 JAS46588.1 GECU01005294 JAT02413.1 GL763868 EFZ19235.1 GECU01021920 JAS85786.1 AAAB01008984 EAA15067.5 KK853095 KDR11489.1 JXUM01065414 KQ562346 KXJ76104.1 CH477982 EAT34618.1 KK107024 QOIP01000004 EZA62257.1 RLU23434.1 KQ982450 KYQ56272.1 JXUM01052463 KQ561734 KXJ77693.1 DS233123 EDS28269.1 NOWV01000017 RDD45296.1 GFDF01001105 JAV12979.1 GDIQ01154530 JAK97195.1 GDIQ01206924 LRGB01001036 JAK44801.1 KZS13678.1 DS985242 EDV28214.1 KQ979709 KYN19445.1 ATLV01014226 KE524948 KFB38482.1 GFDL01009467 JAV25578.1 DS235809 EEB17327.1 PNF29177.1 ADMH02002194 ETN57883.1 JRES01000410 KNC31486.1 CCAG010014451 EZ422149 ADD18344.1 AJVK01008353 GECL01001728 JAP04396.1 GGMS01011083 MBY80286.1 CH940649 KRF81032.1 CH963857 EDW76052.2 KK856248 PTY25293.1 GDRN01072657 JAI63524.1 GFTR01004587 JAW11839.1 LNIX01000001 OXA64821.1 JXJN01021048 GFXV01000961 MBW12766.1 KQ434796 KZC05799.1 KRF81030.1 CH480818 EDW52440.1 GBYB01006172 JAG75939.1 PYGN01001820 PSN32658.1 GDIQ01009739 JAN84998.1 GDIP01016931 JAM86784.1 GDKW01002612 JAI53983.1 GAHY01000327 JAA77183.1 KQ414615 KOC68671.1 KRF81031.1 BT133007 AEX66193.1 GG666459 EEN68923.1 AE014134 AAF52949.2 AMQN01008410 KB303059 ELU03517.1 GEDC01002557 JAS34741.1 CH933807 EDW13354.2 GBGD01002408 JAC86481.1
KQ459597 KPI94980.1 ODYU01005295 SOQ46014.1 KQ971351 KYB26774.1 GEZM01058144 JAV71867.1 LJIG01016105 KRT81599.1 KQ435711 KOX79625.1 KZ288248 PBC31131.1 NEVH01013258 PNF29178.1 ADTU01006535 KQ762882 OAD55381.1 APGK01035263 KB740923 ENN78234.1 KQ981852 KYN34973.1 KB632333 ERL92669.1 NNAY01002575 OXU21017.1 KQ976538 KYM81320.1 APCN01003428 GL888450 EGI60726.1 KQ977305 KYN03679.1 AXCN02000624 GEBQ01013118 JAT26859.1 GL449511 EFN82565.1 GECU01021708 JAS85998.1 GL443403 EFN62138.1 GECZ01023181 JAS46588.1 GECU01005294 JAT02413.1 GL763868 EFZ19235.1 GECU01021920 JAS85786.1 AAAB01008984 EAA15067.5 KK853095 KDR11489.1 JXUM01065414 KQ562346 KXJ76104.1 CH477982 EAT34618.1 KK107024 QOIP01000004 EZA62257.1 RLU23434.1 KQ982450 KYQ56272.1 JXUM01052463 KQ561734 KXJ77693.1 DS233123 EDS28269.1 NOWV01000017 RDD45296.1 GFDF01001105 JAV12979.1 GDIQ01154530 JAK97195.1 GDIQ01206924 LRGB01001036 JAK44801.1 KZS13678.1 DS985242 EDV28214.1 KQ979709 KYN19445.1 ATLV01014226 KE524948 KFB38482.1 GFDL01009467 JAV25578.1 DS235809 EEB17327.1 PNF29177.1 ADMH02002194 ETN57883.1 JRES01000410 KNC31486.1 CCAG010014451 EZ422149 ADD18344.1 AJVK01008353 GECL01001728 JAP04396.1 GGMS01011083 MBY80286.1 CH940649 KRF81032.1 CH963857 EDW76052.2 KK856248 PTY25293.1 GDRN01072657 JAI63524.1 GFTR01004587 JAW11839.1 LNIX01000001 OXA64821.1 JXJN01021048 GFXV01000961 MBW12766.1 KQ434796 KZC05799.1 KRF81030.1 CH480818 EDW52440.1 GBYB01006172 JAG75939.1 PYGN01001820 PSN32658.1 GDIQ01009739 JAN84998.1 GDIP01016931 JAM86784.1 GDKW01002612 JAI53983.1 GAHY01000327 JAA77183.1 KQ414615 KOC68671.1 KRF81031.1 BT133007 AEX66193.1 GG666459 EEN68923.1 AE014134 AAF52949.2 AMQN01008410 KB303059 ELU03517.1 GEDC01002557 JAS34741.1 CH933807 EDW13354.2 GBGD01002408 JAC86481.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000007266
UP000053105
UP000242457
+ More
UP000235965 UP000002358 UP000005205 UP000019118 UP000078541 UP000030742 UP000215335 UP000075900 UP000078540 UP000075881 UP000075840 UP000007755 UP000078542 UP000075886 UP000075884 UP000075901 UP000008237 UP000075903 UP000075902 UP000000311 UP000075882 UP000075880 UP000007062 UP000027135 UP000069940 UP000249989 UP000008820 UP000053097 UP000279307 UP000075809 UP000075920 UP000002320 UP000253843 UP000076858 UP000009022 UP000078492 UP000030765 UP000192221 UP000091820 UP000009046 UP000095301 UP000078200 UP000000673 UP000095300 UP000037069 UP000092444 UP000092462 UP000008792 UP000007798 UP000198287 UP000092460 UP000076407 UP000076502 UP000001292 UP000192223 UP000245037 UP000053825 UP000001554 UP000000803 UP000014760 UP000009192
UP000235965 UP000002358 UP000005205 UP000019118 UP000078541 UP000030742 UP000215335 UP000075900 UP000078540 UP000075881 UP000075840 UP000007755 UP000078542 UP000075886 UP000075884 UP000075901 UP000008237 UP000075903 UP000075902 UP000000311 UP000075882 UP000075880 UP000007062 UP000027135 UP000069940 UP000249989 UP000008820 UP000053097 UP000279307 UP000075809 UP000075920 UP000002320 UP000253843 UP000076858 UP000009022 UP000078492 UP000030765 UP000192221 UP000091820 UP000009046 UP000095301 UP000078200 UP000000673 UP000095300 UP000037069 UP000092444 UP000092462 UP000008792 UP000007798 UP000198287 UP000092460 UP000076407 UP000076502 UP000001292 UP000192223 UP000245037 UP000053825 UP000001554 UP000000803 UP000014760 UP000009192
PRIDE
Interpro
SUPFAM
SSF55920
SSF55920
CDD
ProteinModelPortal
A0A194QT48
A0A2W1BIT7
A0A2A4J6L5
A0A194PV21
A0A2H1W0H7
A0A139WFT0
+ More
A0A1Y1LLJ2 A0A0T6B2D2 A0A0M9A8W1 A0A2A3EJC5 A0A2J7QKU0 K7IPQ7 A0A158P1G2 A0A310SL33 N6TD21 A0A195F3D6 U4UFB3 A0A232ERQ6 A0A182RLC3 A0A151I2D3 A0A182K2U9 A0A182I7M7 F4WYJ4 A0A195CU66 A0A182QAV1 A0A182NBE8 A0A182SVX6 A0A1B6LTB6 E2BNX3 A0A182VE41 A0A1B6IGF3 A0A182U821 E2AWR2 A0A182LMD7 A0A1B6F8M9 A0A1B6JTD1 E9IJM3 A0A182ISS7 A0A1B6IFU2 Q7PWC9 A0A067QPD7 A0A182GI67 Q16K07 A0A026X1V3 A0A151X7C0 A0A182GBE7 A0A182VU20 B0XHA3 A0A369SEK8 A0A1L8E2W7 A0A0P5P536 A0A0P5J527 B3RPM1 A0A195E2Q4 A0A084VKI8 A0A1W4UT93 A0A1A9W0P3 A0A1Q3FDL9 E0VVC1 A0A1I8NDJ9 A0A1A9UDC1 A0A2J7QKR7 W5J3I3 A0A1I8PHN9 A0A0L0CGW9 A0A1B0FEQ1 D3TL49 A0A1B0DPY8 A0A0V0G995 A0A2S2QR89 A0A0Q9W8A1 B4MUC6 A0A2R7X0H5 A0A0P4WD60 A0A224XV30 A0A226F6J1 A0A1B0BV25 A0A182WXR4 A0A2H8TF84 A0A154P1I2 A0A0Q9W9E6 B4HWP5 A0A0C9QZP1 A0A1W4WW54 A0A2P8XKX7 A0A0P6JSW7 A0A0P6BVH9 A0A0P4VWL1 R4G8V2 A0A0L7RD34 A0A0Q9WFK2 H1UUC3 A0A1W4WKA7 C3XSK2 Q9VKV9 R7UJT7 A0A1B6E9Y8 B4KHR7 A0A069DRZ4
A0A1Y1LLJ2 A0A0T6B2D2 A0A0M9A8W1 A0A2A3EJC5 A0A2J7QKU0 K7IPQ7 A0A158P1G2 A0A310SL33 N6TD21 A0A195F3D6 U4UFB3 A0A232ERQ6 A0A182RLC3 A0A151I2D3 A0A182K2U9 A0A182I7M7 F4WYJ4 A0A195CU66 A0A182QAV1 A0A182NBE8 A0A182SVX6 A0A1B6LTB6 E2BNX3 A0A182VE41 A0A1B6IGF3 A0A182U821 E2AWR2 A0A182LMD7 A0A1B6F8M9 A0A1B6JTD1 E9IJM3 A0A182ISS7 A0A1B6IFU2 Q7PWC9 A0A067QPD7 A0A182GI67 Q16K07 A0A026X1V3 A0A151X7C0 A0A182GBE7 A0A182VU20 B0XHA3 A0A369SEK8 A0A1L8E2W7 A0A0P5P536 A0A0P5J527 B3RPM1 A0A195E2Q4 A0A084VKI8 A0A1W4UT93 A0A1A9W0P3 A0A1Q3FDL9 E0VVC1 A0A1I8NDJ9 A0A1A9UDC1 A0A2J7QKR7 W5J3I3 A0A1I8PHN9 A0A0L0CGW9 A0A1B0FEQ1 D3TL49 A0A1B0DPY8 A0A0V0G995 A0A2S2QR89 A0A0Q9W8A1 B4MUC6 A0A2R7X0H5 A0A0P4WD60 A0A224XV30 A0A226F6J1 A0A1B0BV25 A0A182WXR4 A0A2H8TF84 A0A154P1I2 A0A0Q9W9E6 B4HWP5 A0A0C9QZP1 A0A1W4WW54 A0A2P8XKX7 A0A0P6JSW7 A0A0P6BVH9 A0A0P4VWL1 R4G8V2 A0A0L7RD34 A0A0Q9WFK2 H1UUC3 A0A1W4WKA7 C3XSK2 Q9VKV9 R7UJT7 A0A1B6E9Y8 B4KHR7 A0A069DRZ4
PDB
4IU6
E-value=4.50264e-71,
Score=679
Ontologies
GO
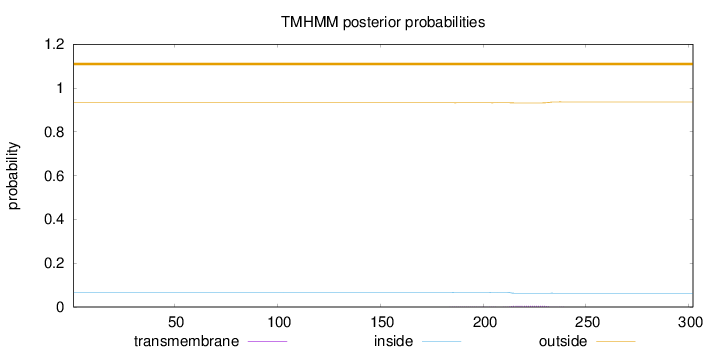
Topology
Length:
302
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1113
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.06720
outside
1 - 302
Population Genetic Test Statistics
Pi
178.601502
Theta
138.648978
Tajima's D
0.741996
CLR
0.04236
CSRT
0.584770761461927
Interpretation
Uncertain
