Gene
KWMTBOMO02855
Annotation
PREDICTED:_protein_N-terminal_glutamine_amidohydrolase_[Amyelois_transitella]
Full name
Protein N-terminal glutamine amidohydrolase
Alternative Name
Protein NH2-terminal glutamine deamidase
Protein tungus
Protein tungus
Location in the cell
Nuclear Reliability : 2.812
Sequence
CDS
ATGCATTTAAACAGCAATGCAAATATTATAAAGAGTGATAAAACTATAATAAAACCTTTATTCCCTAAACGTGAGGAATGTTCTCATGTGTCCTGTTATTGTGAGGAAAACGTTTGGAAACTTTGCCAAGACGTTTCATTGAGAGTACCCGAGGAATTGGATAGATGCTATGTCGTTTTCATTTCGAACCCTTGCCGCACAGTGCCTCTTTGGAAACAAAGGGCCGGGCGTGAAGAAGATCGTCTCGTCATATGGGATTATCATGTGATATTCCTGTATTCTGTGGATATCAAGTCTTGTCTTGTATATGATCTTGATTCAGAATTACCATTTCCAACATTTTTCCACAAATATGTAACAGAAACTTTTCGCACAGACCAAGTCCTCAAATCAGATTTCCACAGGTTCTTTCGTGTTGTGACTGCAAAACAATTTCTGCAACTTTTTTCATCAGATAGACGTCATATGAAAAGACCAGATGGTTCATGGATTAAACCCCCACCACCCTACCCAGCAATAAGCACTTCAGTCTCAACACACAACTTGGATGAGTACATCAATATGGATGTAGACACAGGACCTGGGCAGGTCTACAACTTGACAGATTTTGTCCAACGTTTCTACAAAATTAACAAGTAG
Protein
MHLNSNANIIKSDKTIIKPLFPKREECSHVSCYCEENVWKLCQDVSLRVPEELDRCYVVFISNPCRTVPLWKQRAGREEDRLVIWDYHVIFLYSVDIKSCLVYDLDSELPFPTFFHKYVTETFRTDQVLKSDFHRFFRVVTAKQFLQLFSSDRRHMKRPDGSWIKPPPPYPAISTSVSTHNLDEYINMDVDTGPGQVYNLTDFVQRFYKINK
Summary
Description
Mediates the side-chain deamidation of N-terminal glutamine residues to glutamate, an important step in N-end rule pathway of protein degradation. Conversion of the resulting N-terminal glutamine to glutamate renders the protein susceptible to arginylation, polyubiquitination and degradation as specified by the N-end rule. Does not act on substrates with internal or C-terminal glutamine and does not act on non-glutamine residues in any position.
Catalytic Activity
H2O + N-terminal L-glutaminyl-[protein] = N-terminal L-glutamyl-[protein] + NH4(+)
Subunit
Monomer.
Similarity
Belongs to the NTAQ1 family.
Keywords
Complete proteome
Hydrolase
Reference proteome
Feature
chain Protein N-terminal glutamine amidohydrolase
Uniprot
A0A2A4J6B1
A0A2H1VYX9
A0A194QTR1
A0A212EK23
A0A1E1W9Q8
A0A194PNH3
+ More
D6WRJ5 A0A1Y1KY22 A0A067QP63 H9J1Z1 A0A1W4ULI2 Q7Q968 B4J8A0 A0A084WJ98 A0A0J9RDF7 B4P6V4 B4QGZ1 B4HSF8 B3NQ86 A0A0B4K882 Q7K2Y9 W8BW00 A0A034VK98 A0A0K8UUZ4 A0A0Q9X7D5 B4KPY6 A0A0A1WH88 B4MQL4 A0A0Q9W0P2 B4MDT2 B3MCF3 A0A1I8NU78 A0A0M3QUL4 Q28WL0 A0A1I8ML72 A0A3B0IXX0 A0A1J1HZC4 B4H701 A0A026WLJ5 E0VBN0 A0A232F4M8 K7IQY2 A0A182WS28 A0A182RQE3 A0A195BJD2 A0A158NQ10 A0A195DFQ1 A0A195CVY2 F4WT76 A0A195FNE7 A0A182WML3 A0A151XG06 A0A182SFM4 A0A182JWU6 A0A182NNL3 W5JMZ8 E2AWL5 A0A0T6BA81 A0A182FEB1 A0A182IK86 A0A088AA24 A0A0L7R6U5 A0A1L8DA99 Q173D8 A0A336MTL6 A0A2A3EIU9 V9IBJ3 A0A1A9VPZ4 A0A0C9RD30 A0A1A9X3U0 A0A182QQ51 A0A182LHY3 A0A182HXS3 A0A182VHF6 A0A182TN87 D3PGL7 A0A0P4VPD3 A0A182PV43 A0A154P5A8 A0A182LSF0 A0A336M3I3 A0A0M9AAF9 E2BVQ0 A0A1B0ES12 B0X1U8 A0A0C9QVY8 A0A226EII0 A0A2R5LAZ0 A0A1A9ZNF1 A0A1B0CN57 A0A293L7T8 A0A1B6JUJ8 A0A1Z5L9H9 A0A1E1X6B2 A0A1B6G201 A0A1B6GML7 A0A3B4BUU8 G3MR17 A0A3P8TNN6
D6WRJ5 A0A1Y1KY22 A0A067QP63 H9J1Z1 A0A1W4ULI2 Q7Q968 B4J8A0 A0A084WJ98 A0A0J9RDF7 B4P6V4 B4QGZ1 B4HSF8 B3NQ86 A0A0B4K882 Q7K2Y9 W8BW00 A0A034VK98 A0A0K8UUZ4 A0A0Q9X7D5 B4KPY6 A0A0A1WH88 B4MQL4 A0A0Q9W0P2 B4MDT2 B3MCF3 A0A1I8NU78 A0A0M3QUL4 Q28WL0 A0A1I8ML72 A0A3B0IXX0 A0A1J1HZC4 B4H701 A0A026WLJ5 E0VBN0 A0A232F4M8 K7IQY2 A0A182WS28 A0A182RQE3 A0A195BJD2 A0A158NQ10 A0A195DFQ1 A0A195CVY2 F4WT76 A0A195FNE7 A0A182WML3 A0A151XG06 A0A182SFM4 A0A182JWU6 A0A182NNL3 W5JMZ8 E2AWL5 A0A0T6BA81 A0A182FEB1 A0A182IK86 A0A088AA24 A0A0L7R6U5 A0A1L8DA99 Q173D8 A0A336MTL6 A0A2A3EIU9 V9IBJ3 A0A1A9VPZ4 A0A0C9RD30 A0A1A9X3U0 A0A182QQ51 A0A182LHY3 A0A182HXS3 A0A182VHF6 A0A182TN87 D3PGL7 A0A0P4VPD3 A0A182PV43 A0A154P5A8 A0A182LSF0 A0A336M3I3 A0A0M9AAF9 E2BVQ0 A0A1B0ES12 B0X1U8 A0A0C9QVY8 A0A226EII0 A0A2R5LAZ0 A0A1A9ZNF1 A0A1B0CN57 A0A293L7T8 A0A1B6JUJ8 A0A1Z5L9H9 A0A1E1X6B2 A0A1B6G201 A0A1B6GML7 A0A3B4BUU8 G3MR17 A0A3P8TNN6
EC Number
3.5.1.122
Pubmed
26354079
22118469
18362917
19820115
28004739
24845553
+ More
19121390 12364791 17994087 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 12593794 24495485 25348373 25830018 18057021 15632085 25315136 24508170 30249741 20566863 28648823 20075255 21347285 21719571 20920257 23761445 20798317 17510324 20966253 28528879 28503490 22216098
19121390 12364791 17994087 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 12593794 24495485 25348373 25830018 18057021 15632085 25315136 24508170 30249741 20566863 28648823 20075255 21347285 21719571 20920257 23761445 20798317 17510324 20966253 28528879 28503490 22216098
EMBL
NWSH01003044
PCG67064.1
ODYU01005295
SOQ46013.1
KQ461150
KPJ08699.1
+ More
AGBW02014356 OWR41824.1 GDQN01007341 JAT83713.1 KQ459597 KPI94981.1 KQ971351 EFA06576.1 GEZM01072779 JAV65401.1 KK853098 KDR11405.1 BABH01007715 AAAB01008905 CH916367 ATLV01024001 ATLV01024002 KE525348 KFB50292.1 CM002911 KMY94093.1 CM000158 CM000362 CH480816 CH954179 AE013599 AFH08105.1 AFH08106.1 AHN56265.1 AY060439 GAMC01003128 JAC03428.1 GAKP01016028 JAC42924.1 GDHF01022149 GDHF01017573 GDHF01013676 GDHF01000109 JAI30165.1 JAI34741.1 JAI38638.1 JAI52205.1 CH933808 KRG03893.1 GBXI01015898 GBXI01012613 GBXI01005499 JAC98393.1 JAD01679.1 JAD08793.1 CH963849 CH940662 KRF78406.1 KRF78407.1 CH902619 CP012524 ALC40833.1 CM000071 OUUW01000001 SPP72724.1 CVRI01000035 CRK92676.1 CH479216 EDW33637.1 KK107154 QOIP01000001 EZA56915.1 RLU26313.1 DS235033 EEB10786.1 NNAY01001025 OXU25439.1 KQ976464 KYM84479.1 ADTU01022847 KQ980934 KYN11294.1 KQ977279 KYN04314.1 GL888332 EGI62605.1 KQ981382 KYN42090.1 KQ982182 KYQ59245.1 ADMH02000584 ETN65752.1 GL443346 EFN62179.1 LJIG01002696 KRT84244.1 KQ414646 KOC66476.1 GFDF01010693 JAV03391.1 CH477426 EAT41148.1 UFQT01002270 SSX33055.1 KZ288249 PBC31116.1 JR037861 JR037862 AEY58002.1 AEY58003.1 GBYB01004811 JAG74578.1 AXCN02001920 APCN01005343 BT120773 ADD24413.1 GDRN01124910 JAI56693.1 KQ434822 KZC07116.1 AXCM01018848 UFQS01000501 UFQT01000501 SSX04448.1 SSX24812.1 KQ435700 KOX80535.1 GL450924 EFN80233.1 CCAG010023106 DS232270 EDS38826.1 GBYB01004810 GBYB01004812 JAG74577.1 JAG74579.1 LNIX01000004 OXA56356.1 GGLE01002565 MBY06691.1 AJWK01019817 GFWV01004077 MAA28807.1 GECU01036938 GECU01017668 GECU01004818 JAS70768.1 JAS90038.1 JAT02889.1 GFJQ02003359 JAW03611.1 GFAC01004398 JAT94790.1 GECZ01013358 JAS56411.1 GECZ01017225 GECZ01006112 JAS52544.1 JAS63657.1 JO844318 AEO35935.1
AGBW02014356 OWR41824.1 GDQN01007341 JAT83713.1 KQ459597 KPI94981.1 KQ971351 EFA06576.1 GEZM01072779 JAV65401.1 KK853098 KDR11405.1 BABH01007715 AAAB01008905 CH916367 ATLV01024001 ATLV01024002 KE525348 KFB50292.1 CM002911 KMY94093.1 CM000158 CM000362 CH480816 CH954179 AE013599 AFH08105.1 AFH08106.1 AHN56265.1 AY060439 GAMC01003128 JAC03428.1 GAKP01016028 JAC42924.1 GDHF01022149 GDHF01017573 GDHF01013676 GDHF01000109 JAI30165.1 JAI34741.1 JAI38638.1 JAI52205.1 CH933808 KRG03893.1 GBXI01015898 GBXI01012613 GBXI01005499 JAC98393.1 JAD01679.1 JAD08793.1 CH963849 CH940662 KRF78406.1 KRF78407.1 CH902619 CP012524 ALC40833.1 CM000071 OUUW01000001 SPP72724.1 CVRI01000035 CRK92676.1 CH479216 EDW33637.1 KK107154 QOIP01000001 EZA56915.1 RLU26313.1 DS235033 EEB10786.1 NNAY01001025 OXU25439.1 KQ976464 KYM84479.1 ADTU01022847 KQ980934 KYN11294.1 KQ977279 KYN04314.1 GL888332 EGI62605.1 KQ981382 KYN42090.1 KQ982182 KYQ59245.1 ADMH02000584 ETN65752.1 GL443346 EFN62179.1 LJIG01002696 KRT84244.1 KQ414646 KOC66476.1 GFDF01010693 JAV03391.1 CH477426 EAT41148.1 UFQT01002270 SSX33055.1 KZ288249 PBC31116.1 JR037861 JR037862 AEY58002.1 AEY58003.1 GBYB01004811 JAG74578.1 AXCN02001920 APCN01005343 BT120773 ADD24413.1 GDRN01124910 JAI56693.1 KQ434822 KZC07116.1 AXCM01018848 UFQS01000501 UFQT01000501 SSX04448.1 SSX24812.1 KQ435700 KOX80535.1 GL450924 EFN80233.1 CCAG010023106 DS232270 EDS38826.1 GBYB01004810 GBYB01004812 JAG74577.1 JAG74579.1 LNIX01000004 OXA56356.1 GGLE01002565 MBY06691.1 AJWK01019817 GFWV01004077 MAA28807.1 GECU01036938 GECU01017668 GECU01004818 JAS70768.1 JAS90038.1 JAT02889.1 GFJQ02003359 JAW03611.1 GFAC01004398 JAT94790.1 GECZ01013358 JAS56411.1 GECZ01017225 GECZ01006112 JAS52544.1 JAS63657.1 JO844318 AEO35935.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000007266
UP000027135
+ More
UP000005204 UP000192221 UP000007062 UP000001070 UP000030765 UP000002282 UP000000304 UP000001292 UP000008711 UP000000803 UP000009192 UP000007798 UP000008792 UP000007801 UP000095300 UP000092553 UP000001819 UP000095301 UP000268350 UP000183832 UP000008744 UP000053097 UP000279307 UP000009046 UP000215335 UP000002358 UP000076407 UP000075900 UP000078540 UP000005205 UP000078492 UP000078542 UP000007755 UP000078541 UP000075920 UP000075809 UP000075901 UP000075881 UP000075884 UP000000673 UP000000311 UP000069272 UP000075880 UP000005203 UP000053825 UP000008820 UP000242457 UP000078200 UP000091820 UP000075886 UP000075882 UP000075840 UP000075903 UP000075902 UP000075885 UP000076502 UP000075883 UP000053105 UP000008237 UP000092444 UP000002320 UP000198287 UP000092445 UP000092461 UP000261440 UP000265080
UP000005204 UP000192221 UP000007062 UP000001070 UP000030765 UP000002282 UP000000304 UP000001292 UP000008711 UP000000803 UP000009192 UP000007798 UP000008792 UP000007801 UP000095300 UP000092553 UP000001819 UP000095301 UP000268350 UP000183832 UP000008744 UP000053097 UP000279307 UP000009046 UP000215335 UP000002358 UP000076407 UP000075900 UP000078540 UP000005205 UP000078492 UP000078542 UP000007755 UP000078541 UP000075920 UP000075809 UP000075901 UP000075881 UP000075884 UP000000673 UP000000311 UP000069272 UP000075880 UP000005203 UP000053825 UP000008820 UP000242457 UP000078200 UP000091820 UP000075886 UP000075882 UP000075840 UP000075903 UP000075902 UP000075885 UP000076502 UP000075883 UP000053105 UP000008237 UP000092444 UP000002320 UP000198287 UP000092445 UP000092461 UP000261440 UP000265080
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J6B1
A0A2H1VYX9
A0A194QTR1
A0A212EK23
A0A1E1W9Q8
A0A194PNH3
+ More
D6WRJ5 A0A1Y1KY22 A0A067QP63 H9J1Z1 A0A1W4ULI2 Q7Q968 B4J8A0 A0A084WJ98 A0A0J9RDF7 B4P6V4 B4QGZ1 B4HSF8 B3NQ86 A0A0B4K882 Q7K2Y9 W8BW00 A0A034VK98 A0A0K8UUZ4 A0A0Q9X7D5 B4KPY6 A0A0A1WH88 B4MQL4 A0A0Q9W0P2 B4MDT2 B3MCF3 A0A1I8NU78 A0A0M3QUL4 Q28WL0 A0A1I8ML72 A0A3B0IXX0 A0A1J1HZC4 B4H701 A0A026WLJ5 E0VBN0 A0A232F4M8 K7IQY2 A0A182WS28 A0A182RQE3 A0A195BJD2 A0A158NQ10 A0A195DFQ1 A0A195CVY2 F4WT76 A0A195FNE7 A0A182WML3 A0A151XG06 A0A182SFM4 A0A182JWU6 A0A182NNL3 W5JMZ8 E2AWL5 A0A0T6BA81 A0A182FEB1 A0A182IK86 A0A088AA24 A0A0L7R6U5 A0A1L8DA99 Q173D8 A0A336MTL6 A0A2A3EIU9 V9IBJ3 A0A1A9VPZ4 A0A0C9RD30 A0A1A9X3U0 A0A182QQ51 A0A182LHY3 A0A182HXS3 A0A182VHF6 A0A182TN87 D3PGL7 A0A0P4VPD3 A0A182PV43 A0A154P5A8 A0A182LSF0 A0A336M3I3 A0A0M9AAF9 E2BVQ0 A0A1B0ES12 B0X1U8 A0A0C9QVY8 A0A226EII0 A0A2R5LAZ0 A0A1A9ZNF1 A0A1B0CN57 A0A293L7T8 A0A1B6JUJ8 A0A1Z5L9H9 A0A1E1X6B2 A0A1B6G201 A0A1B6GML7 A0A3B4BUU8 G3MR17 A0A3P8TNN6
D6WRJ5 A0A1Y1KY22 A0A067QP63 H9J1Z1 A0A1W4ULI2 Q7Q968 B4J8A0 A0A084WJ98 A0A0J9RDF7 B4P6V4 B4QGZ1 B4HSF8 B3NQ86 A0A0B4K882 Q7K2Y9 W8BW00 A0A034VK98 A0A0K8UUZ4 A0A0Q9X7D5 B4KPY6 A0A0A1WH88 B4MQL4 A0A0Q9W0P2 B4MDT2 B3MCF3 A0A1I8NU78 A0A0M3QUL4 Q28WL0 A0A1I8ML72 A0A3B0IXX0 A0A1J1HZC4 B4H701 A0A026WLJ5 E0VBN0 A0A232F4M8 K7IQY2 A0A182WS28 A0A182RQE3 A0A195BJD2 A0A158NQ10 A0A195DFQ1 A0A195CVY2 F4WT76 A0A195FNE7 A0A182WML3 A0A151XG06 A0A182SFM4 A0A182JWU6 A0A182NNL3 W5JMZ8 E2AWL5 A0A0T6BA81 A0A182FEB1 A0A182IK86 A0A088AA24 A0A0L7R6U5 A0A1L8DA99 Q173D8 A0A336MTL6 A0A2A3EIU9 V9IBJ3 A0A1A9VPZ4 A0A0C9RD30 A0A1A9X3U0 A0A182QQ51 A0A182LHY3 A0A182HXS3 A0A182VHF6 A0A182TN87 D3PGL7 A0A0P4VPD3 A0A182PV43 A0A154P5A8 A0A182LSF0 A0A336M3I3 A0A0M9AAF9 E2BVQ0 A0A1B0ES12 B0X1U8 A0A0C9QVY8 A0A226EII0 A0A2R5LAZ0 A0A1A9ZNF1 A0A1B0CN57 A0A293L7T8 A0A1B6JUJ8 A0A1Z5L9H9 A0A1E1X6B2 A0A1B6G201 A0A1B6GML7 A0A3B4BUU8 G3MR17 A0A3P8TNN6
PDB
4W79
E-value=2.50946e-47,
Score=473
Ontologies
GO
PANTHER
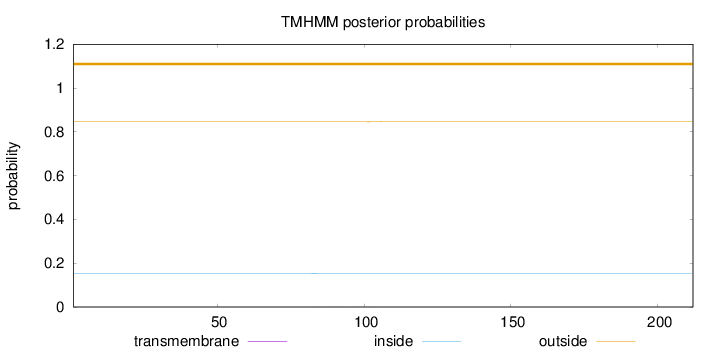
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0206
Exp number, first 60 AAs:
0
Total prob of N-in:
0.15431
outside
1 - 212
Population Genetic Test Statistics
Pi
246.17576
Theta
177.977735
Tajima's D
1.244517
CLR
0
CSRT
0.726813659317034
Interpretation
Possibly Positive selection
