Gene
KWMTBOMO02845 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003522
Annotation
peptidyl-prolyl_cis-trans_isomerase_[Bombyx_mori]
Full name
Peptidyl-prolyl cis-trans isomerase
Alternative Name
Cyclophilin
Cyclosporin A-binding protein
Rotamase
Cyclosporin A-binding protein
Rotamase
Location in the cell
Cytoplasmic Reliability : 2.938
Sequence
CDS
ATGATACCGGTATCAATGCTTCGGCGAACTTTAGTCAATGGGACAATATACTCTGCTGCAGCACTTCGTTTTGCTTCGACTCAACCTGATAAACAAGTTTATTTCGATGTAACTGCTGACGGAGAGCCATTGGGACGTATAGTCATAAAATTGAACACGGATGAAGTACCGAAAACTGCTGAGAACTTTAGAGCTCTGTGTACTGGTGAAAAAGGTTTTGGGTACAAGGGTTCATCATTTCATAGAATAATACCAGACTTCATGTGTCAAGGTGGAGATTTTACCAATCACAACGGTACCGGCGGAAAGTCTATTTACGGGCGTACCTTCTCAGATGAGAATTTCAAATTAAAGCATACTGGGCCTGGAATTTTATCAATGGCGAATGCTGGACCGAATACAAATGGGTCTCAATTTTTTATAACCACAGTTCCGACTCCTTGGTTGGATAGAAAACATGTAGTATTTGGGTCTGTGGTTGAAGGTATGGATGTTGTGCAGAAAATGGAAAAACTTGGTTCTAAAAGTGGTAAACCAATAAAGAAAGTTGTCATCTCAGATTCAGGTGATTATCATGAAGAACTAATTTTGAAGAGTGCAGATTGA
Protein
MIPVSMLRRTLVNGTIYSAAALRFASTQPDKQVYFDVTADGEPLGRIVIKLNTDEVPKTAENFRALCTGEKGFGYKGSSFHRIIPDFMCQGGDFTNHNGTGGKSIYGRTFSDENFKLKHTGPGILSMANAGPNTNGSQFFITTVPTPWLDRKHVVFGSVVEGMDVVQKMEKLGSKSGKPIKKVVISDSGDYHEELILKSAD
Summary
Description
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides.
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Similarity
Belongs to the cyclophilin-type PPIase family.
Belongs to the cyclophilin-type PPIase family. PPIase A subfamily.
Belongs to the cyclophilin-type PPIase family. PPIase A subfamily.
Keywords
Cytoplasm
Isomerase
Rotamase
Complete proteome
Direct protein sequencing
Phosphoprotein
Reference proteome
Feature
chain Peptidyl-prolyl cis-trans isomerase
Uniprot
Q2F624
H9J1Y5
A0A212F1Y8
A0A222AJ14
A0A194QTB5
S4P3Q0
+ More
A0A2H1X2S4 A0A0L7LTJ9 A0A2A4JZ62 A0A2W1BTU8 I4DK44 A0A1Y1L1V2 R4UW88 A0A3B3YI88 A0A1L8EEJ3 A0A1L8E8Y7 A0A1L8EEP6 M3ZRY3 A0A3B5M1H5 A0A023FFI9 A0A3B3UWM9 A0A087XTP2 T1PHF9 A0A1E1X7M1 A0A1D2N7H2 A0A232FLD2 A0A1L8EF36 A0A1Y1L5L4 A0A023FGK4 A0A023GH41 A0A023FXC1 L7M4A5 K7IM71 A0A1Y2HML4 A0A0M4ENB3 N6UJM4 A0A1L8E8G0 B4L5M3 A0A1L8E917 A0A224YFI0 A0A131YUP7 A0A067QY82 A0A2J7RN34 L7WSJ0 D3TMT7 A0A2P8Z1V1 A2I3U7 A0A0K8S607 K7J0P3 A0A0T6AV63 C3Z1U8 A0A131XLA0 A0A026WVJ3 A0A2I4B5Z2 A0A194RNK0 A0A0C9RAF3 A0A034WM88 A0A220XK87 F4WLL7 A0A0K8VZ87 E9IVQ3 A0A0A1XC91 A0A0K8VBA9 A0A1Z5KXX2 B4M328 P54985 A0A3B0K1Q9 C9W1E8 A0A1W4W1Y2 A0A0P6IS29 E2AKH8 A0A1L4AAU0 A0A195CSZ2 A0A3M7QSA4 A0A1A8Q475 A0A1A8LL28 A0A146NVB1 R4FMV8 A0A2A3EAB9 V9IIN8 A0A088A8D4 A0A1H6YZM6 I4DJA1 D3WFS2 A0A2I4CE58 B3N0T9 B4IF57 A0A0S7LK77 A0A3G1T1D0 I4DMT7 A0A195AUN3 A0A151XAA5 A0A1I8PXH6 A0A3P9PYV6 A0A170Z8D8 A0A1B6KQK8 A0A1B6G2X9 A8E774 P25007 A0A1I8PXJ9
A0A2H1X2S4 A0A0L7LTJ9 A0A2A4JZ62 A0A2W1BTU8 I4DK44 A0A1Y1L1V2 R4UW88 A0A3B3YI88 A0A1L8EEJ3 A0A1L8E8Y7 A0A1L8EEP6 M3ZRY3 A0A3B5M1H5 A0A023FFI9 A0A3B3UWM9 A0A087XTP2 T1PHF9 A0A1E1X7M1 A0A1D2N7H2 A0A232FLD2 A0A1L8EF36 A0A1Y1L5L4 A0A023FGK4 A0A023GH41 A0A023FXC1 L7M4A5 K7IM71 A0A1Y2HML4 A0A0M4ENB3 N6UJM4 A0A1L8E8G0 B4L5M3 A0A1L8E917 A0A224YFI0 A0A131YUP7 A0A067QY82 A0A2J7RN34 L7WSJ0 D3TMT7 A0A2P8Z1V1 A2I3U7 A0A0K8S607 K7J0P3 A0A0T6AV63 C3Z1U8 A0A131XLA0 A0A026WVJ3 A0A2I4B5Z2 A0A194RNK0 A0A0C9RAF3 A0A034WM88 A0A220XK87 F4WLL7 A0A0K8VZ87 E9IVQ3 A0A0A1XC91 A0A0K8VBA9 A0A1Z5KXX2 B4M328 P54985 A0A3B0K1Q9 C9W1E8 A0A1W4W1Y2 A0A0P6IS29 E2AKH8 A0A1L4AAU0 A0A195CSZ2 A0A3M7QSA4 A0A1A8Q475 A0A1A8LL28 A0A146NVB1 R4FMV8 A0A2A3EAB9 V9IIN8 A0A088A8D4 A0A1H6YZM6 I4DJA1 D3WFS2 A0A2I4CE58 B3N0T9 B4IF57 A0A0S7LK77 A0A3G1T1D0 I4DMT7 A0A195AUN3 A0A151XAA5 A0A1I8PXH6 A0A3P9PYV6 A0A170Z8D8 A0A1B6KQK8 A0A1B6G2X9 A8E774 P25007 A0A1I8PXJ9
EC Number
5.2.1.8
Pubmed
19121390
22118469
26354079
23622113
26227816
28756777
+ More
22651552 28004739 23542700 25315136 28503490 27289101 28648823 25576852 20075255 23537049 17994087 28797301 26830274 24845553 23326229 20353571 29403074 18563158 28049606 24508170 30249741 25348373 21719571 21282665 25830018 28528879 8529654 20650005 24029695 20798317 27895657 30375419 20147296 10731132 12537572 1707759 8500545 18327897
22651552 28004739 23542700 25315136 28503490 27289101 28648823 25576852 20075255 23537049 17994087 28797301 26830274 24845553 23326229 20353571 29403074 18563158 28049606 24508170 30249741 25348373 21719571 21282665 25830018 28528879 8529654 20650005 24029695 20798317 27895657 30375419 20147296 10731132 12537572 1707759 8500545 18327897
EMBL
DQ311248
ABD36193.1
BABH01007734
AGBW02010786
OWR47746.1
KY563889
+ More
ASO76362.1 KQ461150 KPJ08707.1 GAIX01009397 JAA83163.1 ODYU01013029 SOQ59660.1 JTDY01000112 KOB78790.1 NWSH01000356 PCG77066.1 KZ150007 PZC75163.1 AK401662 KQ459597 BAM18284.1 KPI94971.1 GEZM01066883 JAV67669.1 KC740692 AGM32516.1 GFDG01001688 JAV17111.1 GFDG01003675 JAV15124.1 GFDG01001653 JAV17146.1 GBBK01004617 JAC19865.1 AYCK01021168 KA647328 AFP61957.1 GFAC01003935 JAT95253.1 LJIJ01000175 ODN01025.1 NNAY01000049 OXU31554.1 GFDG01001515 JAV17284.1 GEZM01066882 JAV67670.1 GBBK01004624 JAC19858.1 GBBM01003383 JAC32035.1 GBBL01001965 JAC25355.1 GACK01006164 JAA58870.1 MCFL01000020 ORZ35830.1 CP012526 ALC47077.1 APGK01021421 KB740322 KB631815 ENN80871.1 ERL86427.1 GFDG01003906 JAV14893.1 CH933811 EDW06482.2 GFDG01003700 JAV15099.1 GFPF01004579 MAA15725.1 GEDV01006295 JAP82262.1 KK852834 KDR15281.1 NEVH01002545 PNF42238.1 KC118982 AGC84404.1 EZ422739 ADD19015.1 PYGN01000238 PSN50434.1 PSN50460.1 EF070450 ABM55516.1 GBRD01017243 JAG48584.1 LJIG01022734 KRT78986.1 GG666573 EEN53465.1 GEFH01001641 JAP66940.1 KK107085 QOIP01000008 EZA60013.1 RLU19871.1 KQ459984 KPJ18989.1 GBYB01010057 JAG79824.1 GAKP01003682 JAC55270.1 KY273928 ASL05640.1 GL888208 EGI64953.1 GDHF01008122 JAI44192.1 GL766328 EFZ15350.1 GBXI01005258 JAD09034.1 GDHF01016509 JAI35805.1 GFJQ02007329 JAV99640.1 CH940651 EDW65203.1 X87418 OUUW01000011 SPP86602.1 EZ406096 ACX53895.1 GDIQ01002400 JAN92337.1 GL440281 EFN66029.1 KX268378 API65179.1 KQ977372 KYN03244.1 REGN01005313 RNA13848.1 HAEI01001586 HAEH01009885 SBR88301.1 HAEF01007610 HAEG01007631 SBR45031.1 GCES01150890 JAQ35432.1 GAHY01000813 JAA76697.1 KZ288325 PBC28011.1 JR049570 AEY61003.1 FNYJ01000003 SEJ42832.1 AK401369 KQ459220 BAM17991.1 KPJ02870.1 GU046311 ADC80505.1 CH902644 EDV34818.2 CH480832 EDW46111.1 GBYX01166845 JAO89403.1 MG846936 AXY94788.1 AK402605 BAM19227.1 KQ976738 KYM75891.1 KQ982351 KYQ57250.1 GEMB01002505 JAS00681.1 GEBQ01026251 JAT13726.1 GECZ01012974 JAS56795.1 BT031016 ABV82398.1 AE014298 BT009946 M62398
ASO76362.1 KQ461150 KPJ08707.1 GAIX01009397 JAA83163.1 ODYU01013029 SOQ59660.1 JTDY01000112 KOB78790.1 NWSH01000356 PCG77066.1 KZ150007 PZC75163.1 AK401662 KQ459597 BAM18284.1 KPI94971.1 GEZM01066883 JAV67669.1 KC740692 AGM32516.1 GFDG01001688 JAV17111.1 GFDG01003675 JAV15124.1 GFDG01001653 JAV17146.1 GBBK01004617 JAC19865.1 AYCK01021168 KA647328 AFP61957.1 GFAC01003935 JAT95253.1 LJIJ01000175 ODN01025.1 NNAY01000049 OXU31554.1 GFDG01001515 JAV17284.1 GEZM01066882 JAV67670.1 GBBK01004624 JAC19858.1 GBBM01003383 JAC32035.1 GBBL01001965 JAC25355.1 GACK01006164 JAA58870.1 MCFL01000020 ORZ35830.1 CP012526 ALC47077.1 APGK01021421 KB740322 KB631815 ENN80871.1 ERL86427.1 GFDG01003906 JAV14893.1 CH933811 EDW06482.2 GFDG01003700 JAV15099.1 GFPF01004579 MAA15725.1 GEDV01006295 JAP82262.1 KK852834 KDR15281.1 NEVH01002545 PNF42238.1 KC118982 AGC84404.1 EZ422739 ADD19015.1 PYGN01000238 PSN50434.1 PSN50460.1 EF070450 ABM55516.1 GBRD01017243 JAG48584.1 LJIG01022734 KRT78986.1 GG666573 EEN53465.1 GEFH01001641 JAP66940.1 KK107085 QOIP01000008 EZA60013.1 RLU19871.1 KQ459984 KPJ18989.1 GBYB01010057 JAG79824.1 GAKP01003682 JAC55270.1 KY273928 ASL05640.1 GL888208 EGI64953.1 GDHF01008122 JAI44192.1 GL766328 EFZ15350.1 GBXI01005258 JAD09034.1 GDHF01016509 JAI35805.1 GFJQ02007329 JAV99640.1 CH940651 EDW65203.1 X87418 OUUW01000011 SPP86602.1 EZ406096 ACX53895.1 GDIQ01002400 JAN92337.1 GL440281 EFN66029.1 KX268378 API65179.1 KQ977372 KYN03244.1 REGN01005313 RNA13848.1 HAEI01001586 HAEH01009885 SBR88301.1 HAEF01007610 HAEG01007631 SBR45031.1 GCES01150890 JAQ35432.1 GAHY01000813 JAA76697.1 KZ288325 PBC28011.1 JR049570 AEY61003.1 FNYJ01000003 SEJ42832.1 AK401369 KQ459220 BAM17991.1 KPJ02870.1 GU046311 ADC80505.1 CH902644 EDV34818.2 CH480832 EDW46111.1 GBYX01166845 JAO89403.1 MG846936 AXY94788.1 AK402605 BAM19227.1 KQ976738 KYM75891.1 KQ982351 KYQ57250.1 GEMB01002505 JAS00681.1 GEBQ01026251 JAT13726.1 GECZ01012974 JAS56795.1 BT031016 ABV82398.1 AE014298 BT009946 M62398
Proteomes
UP000005204
UP000007151
UP000053240
UP000037510
UP000218220
UP000053268
+ More
UP000261480 UP000002852 UP000261380 UP000261500 UP000028760 UP000095301 UP000094527 UP000215335 UP000002358 UP000193411 UP000092553 UP000019118 UP000030742 UP000009192 UP000027135 UP000235965 UP000245037 UP000001554 UP000053097 UP000279307 UP000192220 UP000007755 UP000008792 UP000268350 UP000192221 UP000000311 UP000078542 UP000276133 UP000242457 UP000005203 UP000199219 UP000007801 UP000001292 UP000078540 UP000075809 UP000095300 UP000242638 UP000000803
UP000261480 UP000002852 UP000261380 UP000261500 UP000028760 UP000095301 UP000094527 UP000215335 UP000002358 UP000193411 UP000092553 UP000019118 UP000030742 UP000009192 UP000027135 UP000235965 UP000245037 UP000001554 UP000053097 UP000279307 UP000192220 UP000007755 UP000008792 UP000268350 UP000192221 UP000000311 UP000078542 UP000276133 UP000242457 UP000005203 UP000199219 UP000007801 UP000001292 UP000078540 UP000075809 UP000095300 UP000242638 UP000000803
Pfam
PF00160 Pro_isomerase
Interpro
SUPFAM
SSF50891
SSF50891
Gene 3D
ProteinModelPortal
Q2F624
H9J1Y5
A0A212F1Y8
A0A222AJ14
A0A194QTB5
S4P3Q0
+ More
A0A2H1X2S4 A0A0L7LTJ9 A0A2A4JZ62 A0A2W1BTU8 I4DK44 A0A1Y1L1V2 R4UW88 A0A3B3YI88 A0A1L8EEJ3 A0A1L8E8Y7 A0A1L8EEP6 M3ZRY3 A0A3B5M1H5 A0A023FFI9 A0A3B3UWM9 A0A087XTP2 T1PHF9 A0A1E1X7M1 A0A1D2N7H2 A0A232FLD2 A0A1L8EF36 A0A1Y1L5L4 A0A023FGK4 A0A023GH41 A0A023FXC1 L7M4A5 K7IM71 A0A1Y2HML4 A0A0M4ENB3 N6UJM4 A0A1L8E8G0 B4L5M3 A0A1L8E917 A0A224YFI0 A0A131YUP7 A0A067QY82 A0A2J7RN34 L7WSJ0 D3TMT7 A0A2P8Z1V1 A2I3U7 A0A0K8S607 K7J0P3 A0A0T6AV63 C3Z1U8 A0A131XLA0 A0A026WVJ3 A0A2I4B5Z2 A0A194RNK0 A0A0C9RAF3 A0A034WM88 A0A220XK87 F4WLL7 A0A0K8VZ87 E9IVQ3 A0A0A1XC91 A0A0K8VBA9 A0A1Z5KXX2 B4M328 P54985 A0A3B0K1Q9 C9W1E8 A0A1W4W1Y2 A0A0P6IS29 E2AKH8 A0A1L4AAU0 A0A195CSZ2 A0A3M7QSA4 A0A1A8Q475 A0A1A8LL28 A0A146NVB1 R4FMV8 A0A2A3EAB9 V9IIN8 A0A088A8D4 A0A1H6YZM6 I4DJA1 D3WFS2 A0A2I4CE58 B3N0T9 B4IF57 A0A0S7LK77 A0A3G1T1D0 I4DMT7 A0A195AUN3 A0A151XAA5 A0A1I8PXH6 A0A3P9PYV6 A0A170Z8D8 A0A1B6KQK8 A0A1B6G2X9 A8E774 P25007 A0A1I8PXJ9
A0A2H1X2S4 A0A0L7LTJ9 A0A2A4JZ62 A0A2W1BTU8 I4DK44 A0A1Y1L1V2 R4UW88 A0A3B3YI88 A0A1L8EEJ3 A0A1L8E8Y7 A0A1L8EEP6 M3ZRY3 A0A3B5M1H5 A0A023FFI9 A0A3B3UWM9 A0A087XTP2 T1PHF9 A0A1E1X7M1 A0A1D2N7H2 A0A232FLD2 A0A1L8EF36 A0A1Y1L5L4 A0A023FGK4 A0A023GH41 A0A023FXC1 L7M4A5 K7IM71 A0A1Y2HML4 A0A0M4ENB3 N6UJM4 A0A1L8E8G0 B4L5M3 A0A1L8E917 A0A224YFI0 A0A131YUP7 A0A067QY82 A0A2J7RN34 L7WSJ0 D3TMT7 A0A2P8Z1V1 A2I3U7 A0A0K8S607 K7J0P3 A0A0T6AV63 C3Z1U8 A0A131XLA0 A0A026WVJ3 A0A2I4B5Z2 A0A194RNK0 A0A0C9RAF3 A0A034WM88 A0A220XK87 F4WLL7 A0A0K8VZ87 E9IVQ3 A0A0A1XC91 A0A0K8VBA9 A0A1Z5KXX2 B4M328 P54985 A0A3B0K1Q9 C9W1E8 A0A1W4W1Y2 A0A0P6IS29 E2AKH8 A0A1L4AAU0 A0A195CSZ2 A0A3M7QSA4 A0A1A8Q475 A0A1A8LL28 A0A146NVB1 R4FMV8 A0A2A3EAB9 V9IIN8 A0A088A8D4 A0A1H6YZM6 I4DJA1 D3WFS2 A0A2I4CE58 B3N0T9 B4IF57 A0A0S7LK77 A0A3G1T1D0 I4DMT7 A0A195AUN3 A0A151XAA5 A0A1I8PXH6 A0A3P9PYV6 A0A170Z8D8 A0A1B6KQK8 A0A1B6G2X9 A8E774 P25007 A0A1I8PXJ9
PDB
4TOT
E-value=2.85359e-69,
Score=661
Ontologies
GO
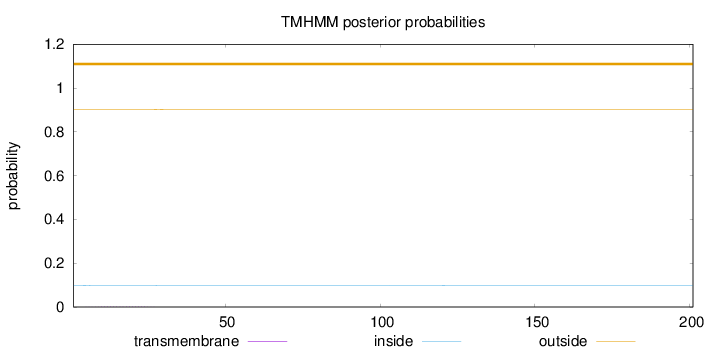
Topology
Subcellular location
Cytoplasm
Length:
201
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02994
Exp number, first 60 AAs:
0.02817
Total prob of N-in:
0.09771
outside
1 - 201
Population Genetic Test Statistics
Pi
171.493425
Theta
150.613604
Tajima's D
0.308434
CLR
0.093179
CSRT
0.459927003649818
Interpretation
Possibly Positive selection
