Gene
KWMTBOMO02831
Pre Gene Modal
BGIBMGA003675
Annotation
PREDICTED:_nucleoside_diphosphate_kinase_7_[Papilio_polytes]
Full name
Nucleoside diphosphate kinase 7
+ More
Nucleoside diphosphate kinase
Nucleoside diphosphate kinase
Alternative Name
nm23-M7
nm23-R7
nm23-H7
nm23-R7
nm23-H7
Location in the cell
Cytoplasmic Reliability : 1.785
Sequence
CDS
ATGGAAGAATCTTACGCTGATAAATATTCTTTCATTGGCGAATGGTATGACAATCAAGCAAATTTAAAACGCCGGTTTAACATTTTTTACTACCCTTCTGATGATACTATAGAAATGTATGACTTAAAAAGTCGAAAAACGTTTGTGAAAAGAGTTAAAGTAAATGGTGTAACTTTAGATAGGTTTTATATTGGTTGTACTTTAAGTATATTAGGTCGTCTTATAAAAATAATTGACTTCGCTTGTGACCATACACGAAAGAAGTTGCACAATGAAATGCAAGTAACTTTTGCTATGATTAAACCCCTTCCAACTGAAATCGTTGGTAAAATTTTAAGTCACTTTCATGAGCATGGTTTGCGGGTAACAAAAATGAAGAAATCACGACTGACTGCTGAAGATATTAATATTTTGTATAGATCTCAGGTCACAGATCCTACATTTCCGTTTCTTTTAGATTACCTCACAGGAGAAATGGTTTATGGTTTAGAGTTAGTTGGAAGAGATGCAGTACTAGTATGTATTAAACTCCTTGGTGATAAAGATCCAAGAAAGGCTGAAATAGGGTCAATAAGAGCTCTCTATGGTACAGATCCTGTTAAAAATTGTGTTCATGCATCAAGCTCATCAGAATCGGCAATAGAGGATATAGAATATTTTTTTCCACCTCCACCTCTAAATTCGACACGACTTTGTCTGACTGCAACTCTCTCGAACTGCACCCTTTGTATAGTAAAGCCTCATGCAATACTAGAGGGAAAACTAGGTGCAGCTTTGGAAGCCATTGATAAGGGTGGTTTCAATGTAACTGCCCTCTACATGCTGATTGTTGAAAATATTAATGCAGCTGAATTTTATGAAGTTTATAAAGGCATTGTGCCTGAATACAAGGGTATGGTTGAAGAATTGTCGAGTGGCCCTTGCGTGGCTATGGAAATAGTAGCGAAAAATAAAAATGTTAACACTGCTGTTGAATTTAGAAAATTAGTAGGTCCTTCTGATCCTGAGATTGCAAGATTACTTCGACCGGATACCCTGAGAGCAAAGTTGGGAAAAACGAAAGTACAAAACGCAGTTCACTGTAGCGATTTAGCAGAAGATGGATTACTAGAAGTTGAATACTTTTTCAAGATCTTAGCATTATAA
Protein
MEESYADKYSFIGEWYDNQANLKRRFNIFYYPSDDTIEMYDLKSRKTFVKRVKVNGVTLDRFYIGCTLSILGRLIKIIDFACDHTRKKLHNEMQVTFAMIKPLPTEIVGKILSHFHEHGLRVTKMKKSRLTAEDINILYRSQVTDPTFPFLLDYLTGEMVYGLELVGRDAVLVCIKLLGDKDPRKAEIGSIRALYGTDPVKNCVHASSSSESAIEDIEYFFPPPPLNSTRLCLTATLSNCTLCIVKPHAILEGKLGAALEAIDKGGFNVTALYMLIVENINAAEFYEVYKGIVPEYKGMVEELSSGPCVAMEIVAKNKNVNTAVEFRKLVGPSDPEIARLLRPDTLRAKLGKTKVQNAVHCSDLAEDGLLEVEYFFKILAL
Summary
Catalytic Activity
a 2'-deoxyribonucleoside 5'-diphosphate + ATP = a 2'-deoxyribonucleoside 5'-triphosphate + ADP
a ribonucleoside 5'-diphosphate + ATP = a ribonucleoside 5'-triphosphate + ADP
a ribonucleoside 5'-diphosphate + ATP = a ribonucleoside 5'-triphosphate + ADP
Cofactor
Mg(2+)
Similarity
Belongs to the NDK family.
Keywords
ATP-binding
Complete proteome
Kinase
Magnesium
Metal-binding
Nucleotide metabolism
Nucleotide-binding
Reference proteome
Transferase
Alternative splicing
Feature
chain Nucleoside diphosphate kinase 7
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2W1BTV9
A0A2A4JZY2
A0A2H1X336
A0A212F1W4
A0A194PNE6
A0A0N1ITX7
+ More
A0A2A3E3V5 A0A087ZWW7 C4B8X0 A0A1B6FBE1 A0A026VWD2 E9J644 A0A1B6LER2 A0A1B6KTD4 A0A2J7QC66 A0A2B4S5Z9 A0A2J7QC52 A0A2J7QC53 A0A151XC28 A0A1S3F4U5 F4X6X1 A0A158NFY0 J9HLV5 R7TZK1 A0A067RKK2 A0A2Y9RKT2 A0A1B6C8C7 A0A0C9QCH4 A7SP47 K9IJ52 A0A2K5ZQK5 A0A096N6Q5 G3VTT6 G7MFA1 E2BWD1 G3TAM2 A0A3P8ZQL6 A0A2K6LGL2 A0A2K5IWR3 Q05BM9 H2MLS7 A0A0D9RF97 A0A2K6BTG8 A0A2K5X5S7 H9FTL1 A0A2K5LKL9 Q80UP4 A0A154P1Y2 C1BYW3 F1LNL9 A0A210R3D6 E2AMH5 A0A3P9LEE9 A0A384CI25 H2ZR10 A0A0L7R3P5 G9KDJ4 A0A195F2D0 Q3UMG6 A0A2K6UUV2 U3E823 A0A2U3Y894 W4Z1L5 W5KX55 G1RY63 Q6PAG9 Q9QXL8 A0A3P8XTR1 F7GQ16 U6CNG2 A0A2K5PVF6 A0A3P9I4A5 H3B3B9 F1PMF2 H0V2N4 H2Q0K0 G1LYF0 Q9QXL7 A0A2J8SAT5 A0A2I2YP06 A0A2R9BXY8 Q9Y5B8 A0A3P8R4P9 A0A384CI35 K9K275 M3WB88 A0A3P9BMP9 A0A3B4XQE1 A0A078A2E8 A0A287AHH1 A0A0P6J402 I3MHE9 I3KT89 A0A2Y9JT67 F6Q3R3 A0A2Y9S527 A0A341B947 G1TFZ9
A0A2A3E3V5 A0A087ZWW7 C4B8X0 A0A1B6FBE1 A0A026VWD2 E9J644 A0A1B6LER2 A0A1B6KTD4 A0A2J7QC66 A0A2B4S5Z9 A0A2J7QC52 A0A2J7QC53 A0A151XC28 A0A1S3F4U5 F4X6X1 A0A158NFY0 J9HLV5 R7TZK1 A0A067RKK2 A0A2Y9RKT2 A0A1B6C8C7 A0A0C9QCH4 A7SP47 K9IJ52 A0A2K5ZQK5 A0A096N6Q5 G3VTT6 G7MFA1 E2BWD1 G3TAM2 A0A3P8ZQL6 A0A2K6LGL2 A0A2K5IWR3 Q05BM9 H2MLS7 A0A0D9RF97 A0A2K6BTG8 A0A2K5X5S7 H9FTL1 A0A2K5LKL9 Q80UP4 A0A154P1Y2 C1BYW3 F1LNL9 A0A210R3D6 E2AMH5 A0A3P9LEE9 A0A384CI25 H2ZR10 A0A0L7R3P5 G9KDJ4 A0A195F2D0 Q3UMG6 A0A2K6UUV2 U3E823 A0A2U3Y894 W4Z1L5 W5KX55 G1RY63 Q6PAG9 Q9QXL8 A0A3P8XTR1 F7GQ16 U6CNG2 A0A2K5PVF6 A0A3P9I4A5 H3B3B9 F1PMF2 H0V2N4 H2Q0K0 G1LYF0 Q9QXL7 A0A2J8SAT5 A0A2I2YP06 A0A2R9BXY8 Q9Y5B8 A0A3P8R4P9 A0A384CI35 K9K275 M3WB88 A0A3P9BMP9 A0A3B4XQE1 A0A078A2E8 A0A287AHH1 A0A0P6J402 I3MHE9 I3KT89 A0A2Y9JT67 F6Q3R3 A0A2Y9S527 A0A341B947 G1TFZ9
EC Number
2.7.4.6
Pubmed
28756777
22118469
26354079
12203911
12481130
15114417
+ More
19527718 24508170 21282665 21719571 21347285 23382650 23254933 24845553 17615350 21709235 22002653 20798317 25069045 15489334 17554307 17431167 25319552 20433749 15057822 28812685 23236062 10349636 11042159 11076861 11217851 12466851 12040188 16141073 19468303 21183079 25243066 25329095 9215903 16341006 21993624 16136131 20010809 20966424 22398555 22722832 14702039 16710414 14654843 21269460 24813606 19892987 17975172 25186727 17495919
19527718 24508170 21282665 21719571 21347285 23382650 23254933 24845553 17615350 21709235 22002653 20798317 25069045 15489334 17554307 17431167 25319552 20433749 15057822 28812685 23236062 10349636 11042159 11076861 11217851 12466851 12040188 16141073 19468303 21183079 25243066 25329095 9215903 16341006 21993624 16136131 20010809 20966424 22398555 22722832 14702039 16710414 14654843 21269460 24813606 19892987 17975172 25186727 17495919
EMBL
KZ150007
PZC75173.1
NWSH01000356
PCG77073.1
ODYU01013029
SOQ59646.1
+ More
AGBW02010786 OWR47735.1 KQ459597 KPI94956.1 KQ435726 KOX78158.1 KZ288388 PBC26395.1 EAAA01000095 AB501294 BAH59279.1 GECZ01022179 JAS47590.1 KK107770 EZA47831.1 GL768221 EFZ11731.1 GEBQ01017802 JAT22175.1 GEBQ01025316 JAT14661.1 NEVH01016292 PNF26162.1 LSMT01000176 PFX24463.1 PNF26163.1 PNF26165.1 KQ982314 KYQ57899.1 GL888818 EGI57903.1 ADTU01014832 AMCR01022026 EJY64649.1 AMQN01010045 KB306898 ELT99378.1 KK852415 KDR24401.1 GEDC01027552 JAS09746.1 GBYB01001004 JAG70771.1 DS469728 EDO34502.1 GABZ01006590 JAA46935.1 AHZZ02017791 AHZZ02017792 AHZZ02017793 AHZZ02017794 AHZZ02017795 AHZZ02017796 AEFK01155615 AEFK01155616 AEFK01155617 AEFK01155618 AEFK01155619 AEFK01155620 AEFK01155621 CM001253 EHH15798.1 GL451103 EFN80035.1 BC038021 AAH38021.1 AQIB01113215 AQIB01113216 AQIB01113217 AQIA01000664 AQIA01000665 AQIA01000666 AQIA01000667 JSUE03003452 JSUE03003453 JSUE03003454 JU334214 JU334215 JU334216 JU334217 JU334218 JU334219 JU475332 AFE77970.1 AFH32136.1 BC049225 AAH49225.1 KQ434786 KZC05140.1 BT079792 ACO14216.1 AABR07021629 NEDP02000661 OWF55446.1 GL440813 EFN65399.1 KQ414663 KOC65507.1 JP014371 AES02969.1 KQ981864 KYN34244.1 AC105161 AK133221 AK144915 CH466520 BAE21565.1 BAE26132.1 EDL39255.1 GAMT01001410 GAMT01001409 GAMT01001408 GAMT01001407 GAMT01001406 GAMT01001405 GAMT01001404 GAMT01001403 GAMS01001375 GAMS01001372 GAMS01001371 GAMS01001370 GAMS01001369 GAMS01001368 GAMR01002945 GAMR01002944 GAMR01002943 GAMR01002942 GAMR01002941 GAMR01002940 GAMR01002939 GAMQ01003950 GAMQ01003949 GAMP01002079 GAMP01002078 GAMP01002077 GAMP01002076 GAMP01002075 JAB10451.1 JAB21761.1 JAB30987.1 JAB37901.1 JAB50676.1 AAGJ04096940 ADFV01184505 ADFV01184506 ADFV01184507 ADFV01184508 ADFV01184509 ADFV01184510 ADFV01184511 ADFV01184512 ADFV01184513 ADFV01184514 BC060314 AAH60314.1 AF202048 HAAF01000294 CCP72120.1 AFYH01047035 AFYH01047036 AFYH01047037 AFYH01047038 AFYH01047039 AFYH01047040 AFYH01047041 AFYH01047042 AFYH01047043 AFYH01047044 AAEX03005219 AAKN02037427 AAKN02037428 AAKN02037429 AAKN02037430 AAKN02037431 AAKN02037432 AACZ04056276 AACZ04056277 AACZ04056278 GABC01000518 GABF01007119 GABD01000341 NBAG03000285 JAA10820.1 JAA15026.1 JAA32759.1 PNI48222.1 ACTA01112981 ACTA01120981 ACTA01128981 ACTA01136981 ACTA01144980 ACTA01152979 ACTA01160978 ACTA01168976 ACTA01176974 ACTA01184972 AF202049 NDHI03003587 PNJ17889.1 CABD030007185 CABD030007186 CABD030007187 CABD030007188 CABD030007189 CABD030007190 CABD030007191 AJFE02086946 AJFE02086947 AF153191 AK094513 AK290701 AL031726 Z99758 AL356852 CH471067 BC006983 JL616087 AEH58294.1 AANG04002195 CCKQ01004544 CDW75698.1 AEMK02000022 GEBF01005558 JAN98074.1 AGTP01015068 AGTP01015069 AERX01011543 AERX01011544 AAGW02007052
AGBW02010786 OWR47735.1 KQ459597 KPI94956.1 KQ435726 KOX78158.1 KZ288388 PBC26395.1 EAAA01000095 AB501294 BAH59279.1 GECZ01022179 JAS47590.1 KK107770 EZA47831.1 GL768221 EFZ11731.1 GEBQ01017802 JAT22175.1 GEBQ01025316 JAT14661.1 NEVH01016292 PNF26162.1 LSMT01000176 PFX24463.1 PNF26163.1 PNF26165.1 KQ982314 KYQ57899.1 GL888818 EGI57903.1 ADTU01014832 AMCR01022026 EJY64649.1 AMQN01010045 KB306898 ELT99378.1 KK852415 KDR24401.1 GEDC01027552 JAS09746.1 GBYB01001004 JAG70771.1 DS469728 EDO34502.1 GABZ01006590 JAA46935.1 AHZZ02017791 AHZZ02017792 AHZZ02017793 AHZZ02017794 AHZZ02017795 AHZZ02017796 AEFK01155615 AEFK01155616 AEFK01155617 AEFK01155618 AEFK01155619 AEFK01155620 AEFK01155621 CM001253 EHH15798.1 GL451103 EFN80035.1 BC038021 AAH38021.1 AQIB01113215 AQIB01113216 AQIB01113217 AQIA01000664 AQIA01000665 AQIA01000666 AQIA01000667 JSUE03003452 JSUE03003453 JSUE03003454 JU334214 JU334215 JU334216 JU334217 JU334218 JU334219 JU475332 AFE77970.1 AFH32136.1 BC049225 AAH49225.1 KQ434786 KZC05140.1 BT079792 ACO14216.1 AABR07021629 NEDP02000661 OWF55446.1 GL440813 EFN65399.1 KQ414663 KOC65507.1 JP014371 AES02969.1 KQ981864 KYN34244.1 AC105161 AK133221 AK144915 CH466520 BAE21565.1 BAE26132.1 EDL39255.1 GAMT01001410 GAMT01001409 GAMT01001408 GAMT01001407 GAMT01001406 GAMT01001405 GAMT01001404 GAMT01001403 GAMS01001375 GAMS01001372 GAMS01001371 GAMS01001370 GAMS01001369 GAMS01001368 GAMR01002945 GAMR01002944 GAMR01002943 GAMR01002942 GAMR01002941 GAMR01002940 GAMR01002939 GAMQ01003950 GAMQ01003949 GAMP01002079 GAMP01002078 GAMP01002077 GAMP01002076 GAMP01002075 JAB10451.1 JAB21761.1 JAB30987.1 JAB37901.1 JAB50676.1 AAGJ04096940 ADFV01184505 ADFV01184506 ADFV01184507 ADFV01184508 ADFV01184509 ADFV01184510 ADFV01184511 ADFV01184512 ADFV01184513 ADFV01184514 BC060314 AAH60314.1 AF202048 HAAF01000294 CCP72120.1 AFYH01047035 AFYH01047036 AFYH01047037 AFYH01047038 AFYH01047039 AFYH01047040 AFYH01047041 AFYH01047042 AFYH01047043 AFYH01047044 AAEX03005219 AAKN02037427 AAKN02037428 AAKN02037429 AAKN02037430 AAKN02037431 AAKN02037432 AACZ04056276 AACZ04056277 AACZ04056278 GABC01000518 GABF01007119 GABD01000341 NBAG03000285 JAA10820.1 JAA15026.1 JAA32759.1 PNI48222.1 ACTA01112981 ACTA01120981 ACTA01128981 ACTA01136981 ACTA01144980 ACTA01152979 ACTA01160978 ACTA01168976 ACTA01176974 ACTA01184972 AF202049 NDHI03003587 PNJ17889.1 CABD030007185 CABD030007186 CABD030007187 CABD030007188 CABD030007189 CABD030007190 CABD030007191 AJFE02086946 AJFE02086947 AF153191 AK094513 AK290701 AL031726 Z99758 AL356852 CH471067 BC006983 JL616087 AEH58294.1 AANG04002195 CCKQ01004544 CDW75698.1 AEMK02000022 GEBF01005558 JAN98074.1 AGTP01015068 AGTP01015069 AERX01011543 AERX01011544 AAGW02007052
Proteomes
UP000218220
UP000007151
UP000053268
UP000053105
UP000242457
UP000005203
+ More
UP000008144 UP000053097 UP000235965 UP000225706 UP000075809 UP000081671 UP000007755 UP000005205 UP000014760 UP000027135 UP000248480 UP000001593 UP000233140 UP000028761 UP000007648 UP000008237 UP000007646 UP000265140 UP000233180 UP000233080 UP000001038 UP000029965 UP000233120 UP000233100 UP000006718 UP000233060 UP000076502 UP000002494 UP000242188 UP000000311 UP000265180 UP000261680 UP000007875 UP000053825 UP000078541 UP000000589 UP000233220 UP000245341 UP000007110 UP000018467 UP000001073 UP000008225 UP000233040 UP000265200 UP000008672 UP000002254 UP000005447 UP000002277 UP000008912 UP000001519 UP000240080 UP000005640 UP000265100 UP000291021 UP000002281 UP000011712 UP000265160 UP000261360 UP000008227 UP000005215 UP000005207 UP000248482 UP000002280 UP000248484 UP000252040 UP000001811
UP000008144 UP000053097 UP000235965 UP000225706 UP000075809 UP000081671 UP000007755 UP000005205 UP000014760 UP000027135 UP000248480 UP000001593 UP000233140 UP000028761 UP000007648 UP000008237 UP000007646 UP000265140 UP000233180 UP000233080 UP000001038 UP000029965 UP000233120 UP000233100 UP000006718 UP000233060 UP000076502 UP000002494 UP000242188 UP000000311 UP000265180 UP000261680 UP000007875 UP000053825 UP000078541 UP000000589 UP000233220 UP000245341 UP000007110 UP000018467 UP000001073 UP000008225 UP000233040 UP000265200 UP000008672 UP000002254 UP000005447 UP000002277 UP000008912 UP000001519 UP000240080 UP000005640 UP000265100 UP000291021 UP000002281 UP000011712 UP000265160 UP000261360 UP000008227 UP000005215 UP000005207 UP000248482 UP000002280 UP000248484 UP000252040 UP000001811
Interpro
SUPFAM
SSF54919
SSF54919
Gene 3D
ProteinModelPortal
A0A2W1BTV9
A0A2A4JZY2
A0A2H1X336
A0A212F1W4
A0A194PNE6
A0A0N1ITX7
+ More
A0A2A3E3V5 A0A087ZWW7 C4B8X0 A0A1B6FBE1 A0A026VWD2 E9J644 A0A1B6LER2 A0A1B6KTD4 A0A2J7QC66 A0A2B4S5Z9 A0A2J7QC52 A0A2J7QC53 A0A151XC28 A0A1S3F4U5 F4X6X1 A0A158NFY0 J9HLV5 R7TZK1 A0A067RKK2 A0A2Y9RKT2 A0A1B6C8C7 A0A0C9QCH4 A7SP47 K9IJ52 A0A2K5ZQK5 A0A096N6Q5 G3VTT6 G7MFA1 E2BWD1 G3TAM2 A0A3P8ZQL6 A0A2K6LGL2 A0A2K5IWR3 Q05BM9 H2MLS7 A0A0D9RF97 A0A2K6BTG8 A0A2K5X5S7 H9FTL1 A0A2K5LKL9 Q80UP4 A0A154P1Y2 C1BYW3 F1LNL9 A0A210R3D6 E2AMH5 A0A3P9LEE9 A0A384CI25 H2ZR10 A0A0L7R3P5 G9KDJ4 A0A195F2D0 Q3UMG6 A0A2K6UUV2 U3E823 A0A2U3Y894 W4Z1L5 W5KX55 G1RY63 Q6PAG9 Q9QXL8 A0A3P8XTR1 F7GQ16 U6CNG2 A0A2K5PVF6 A0A3P9I4A5 H3B3B9 F1PMF2 H0V2N4 H2Q0K0 G1LYF0 Q9QXL7 A0A2J8SAT5 A0A2I2YP06 A0A2R9BXY8 Q9Y5B8 A0A3P8R4P9 A0A384CI35 K9K275 M3WB88 A0A3P9BMP9 A0A3B4XQE1 A0A078A2E8 A0A287AHH1 A0A0P6J402 I3MHE9 I3KT89 A0A2Y9JT67 F6Q3R3 A0A2Y9S527 A0A341B947 G1TFZ9
A0A2A3E3V5 A0A087ZWW7 C4B8X0 A0A1B6FBE1 A0A026VWD2 E9J644 A0A1B6LER2 A0A1B6KTD4 A0A2J7QC66 A0A2B4S5Z9 A0A2J7QC52 A0A2J7QC53 A0A151XC28 A0A1S3F4U5 F4X6X1 A0A158NFY0 J9HLV5 R7TZK1 A0A067RKK2 A0A2Y9RKT2 A0A1B6C8C7 A0A0C9QCH4 A7SP47 K9IJ52 A0A2K5ZQK5 A0A096N6Q5 G3VTT6 G7MFA1 E2BWD1 G3TAM2 A0A3P8ZQL6 A0A2K6LGL2 A0A2K5IWR3 Q05BM9 H2MLS7 A0A0D9RF97 A0A2K6BTG8 A0A2K5X5S7 H9FTL1 A0A2K5LKL9 Q80UP4 A0A154P1Y2 C1BYW3 F1LNL9 A0A210R3D6 E2AMH5 A0A3P9LEE9 A0A384CI25 H2ZR10 A0A0L7R3P5 G9KDJ4 A0A195F2D0 Q3UMG6 A0A2K6UUV2 U3E823 A0A2U3Y894 W4Z1L5 W5KX55 G1RY63 Q6PAG9 Q9QXL8 A0A3P8XTR1 F7GQ16 U6CNG2 A0A2K5PVF6 A0A3P9I4A5 H3B3B9 F1PMF2 H0V2N4 H2Q0K0 G1LYF0 Q9QXL7 A0A2J8SAT5 A0A2I2YP06 A0A2R9BXY8 Q9Y5B8 A0A3P8R4P9 A0A384CI35 K9K275 M3WB88 A0A3P9BMP9 A0A3B4XQE1 A0A078A2E8 A0A287AHH1 A0A0P6J402 I3MHE9 I3KT89 A0A2Y9JT67 F6Q3R3 A0A2Y9S527 A0A341B947 G1TFZ9
PDB
3ZTS
E-value=1.06565e-16,
Score=212
Ontologies
PATHWAY
GO
PANTHER
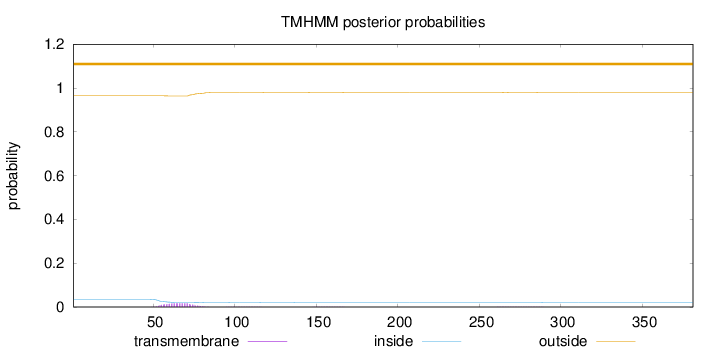
Topology
Length:
381
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.36941
Exp number, first 60 AAs:
0.09001
Total prob of N-in:
0.03505
outside
1 - 381
Population Genetic Test Statistics
Pi
69.108856
Theta
154.606781
Tajima's D
-1.763189
CLR
244.97495
CSRT
0.0300484975751212
Interpretation
Uncertain
