Gene
KWMTBOMO02830
Pre Gene Modal
BGIBMGA003676
Annotation
PREDICTED:_uncharacterized_protein_LOC106717587_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.283
Sequence
CDS
ATGCGACTAATCAGGTCATTTTTCAAAAGAATAACAGATGACTTGAAGCAGAAAAAATTGATTCCATTAAAAATACTGTTCTTCGTTCATGCTTCAACTTTGTACGTGTTGTATCCTTACTTGACGATACATATGCGAGAGCTTGGCATCAATGTTGAGGAAACGGCGATAATGTCTGCCGTAACTCCAGTGGTGTCCATAATAATGCCACCCTTGGCTGGTATGCTTGCAGACAAAATTGGAAATTTTAGACTATTGTTGGCGTTGTCGTCAGTATTGGGCGGTATCTCGGCCTTGTTGTTGTTGGCCGTACCCGTCGGAAGAATAACAATTACATATCCCACAGATGTAGAGCTTTTAACAACCTGTGATGACTCTGCTCTGAGGGTCCGACATATCCAAGAATATCCATGCCTACCCTTGCATCCATATTCATATGACATCAACCTTAGAATTTCATCATGTGGCTTCATTTGTGACGTCCCAGATATAGTTACTCCGAATGAAATTGACAACATATTGCGGAACGACGCATACGATATACACTTGCATTCAAGTTTGGATGCCCAACAACTGGTGTACCGGCACACATTAACTTCATCGGTGACTCGATCGATGACTCCTATGAAAGACCAAAGCACGTCACGAATCCTGAACAATGATCCTTACTTTAATGTAACTGTGTCTAAAATCTCCGACCGAGAGATATTCTTTCCAGCACCAAGACTTTACAAAATGCATTGCTCTCGAGATTTGAATAATGGATCGTGTGTTTTTGGAAGAAATGATATTTTTAAACAAAACGACGAAGGTGGTGAGGAAATGGAAACATTCCGAATGAGAATTTCGCATGATGCACAGAAAACGCCTAAAACTGTAAGAAATTATTGGGTTAGAGCTTTATCTAGTGGAAACGGTTCAATCGACAACGATTACGATGGAGTCGACAGTGTTATATGTAAAAACAGTTATGACGAGGTACTTGACGAAAGTGGAGTTCGAGTGCAAGTCGGATCACGGCAGCTGCTTGGATGCTTAGCTGCTTGTACGGCGATCACATCACGTAATTCTCTTTGTTCAAATACACAACAACCAATTGAGATTGATCCTGCTTTCACATTTTGGACCTACCTTGCTGTGAGAGTGTTTATAGGTATAATAGGCGGTACAGCATTTGCTATGTTTGAGGGTGCAGTGATTGCCATAGTGAGGGATCAAAAAGCGGACTATGGCTTACAGAGAGTCTATGGCAGTATAGGAGGAATGATTTCATCACCTCTTTCTGGACTTTTAATAGACTATGCTAGCAGAGGGAAAGGATACACAGACTTTAGGCCAGCGTTTTATCTTTACGCAATACTAAAAGTCGCTTCCGGTGTATTGATGCTGAGCATCGATCTTGAGTTTAAACAGCCTGCGCAGAACCTCCTCGAAGATGTCATCACAGTATTCAAGAACGTAGAAATCGTTGCGCTGTTCATTGCGTGCTTTATAATGGGCACTGCATGGGGTTACATAGAGAGCTTCTTGTTTTGGCTGCTCCAAGACCTCGGAGCCTCCAGGTCGTTAATGGGGATCACCATAACTGTGGGTGGTGTCGCGGGTCTGCCTCTCCTGGTGCTATCAGGGCCCATCATCAGGAAGTTGGGTCATGCTAATGTATTGTTCATAGGATTTATATTCTACGCCATACGATTGCTTGGCTACTCGCTGATATACAACCCGTGGTTGTGTTTGGTGTTCGAGGCACTGGAGTCGGTAACGTCATCGTTGTCGTTCACCGCAGCGGTGACGTATGCCGCGCGACTCTCCTCCACCACCACCGACACATCCGTACAGGGACTACTTGGTGGCCTGTACTATGGCGTTGGTAAAGGAGCTGGCAGCTTAATAGGCGGATATCTCATGAAGTTCTTCGGCACGAGGCCAACATATCAAATTTTTGCCGGTGCCACTTTAGTCACCGGTTGTATGTACTATCTATTCAATAAATTCTATATTGGAAAGCGACCCGGCAGCGTGAACAACGACATCTGCAAACCGCAGCCAACAGATGTCGAGGGGGTAAAAGGTTCCGACAAAACTTCAAGTCCACTAGATGTCGCTACTGACAAATTTGAGAATGACAAAGTACTAGACTTAAATCACAGAAAGTTAGTAGCGGAATGTACAGATGGCGGTAGTGACTCCGGCGTCGAAAATCCCGCTTACAACGAAACCGAGTGCGCGAACGGATCAAAGAAAGACGAAACGAAATGA
Protein
MRLIRSFFKRITDDLKQKKLIPLKILFFVHASTLYVLYPYLTIHMRELGINVEETAIMSAVTPVVSIIMPPLAGMLADKIGNFRLLLALSSVLGGISALLLLAVPVGRITITYPTDVELLTTCDDSALRVRHIQEYPCLPLHPYSYDINLRISSCGFICDVPDIVTPNEIDNILRNDAYDIHLHSSLDAQQLVYRHTLTSSVTRSMTPMKDQSTSRILNNDPYFNVTVSKISDREIFFPAPRLYKMHCSRDLNNGSCVFGRNDIFKQNDEGGEEMETFRMRISHDAQKTPKTVRNYWVRALSSGNGSIDNDYDGVDSVICKNSYDEVLDESGVRVQVGSRQLLGCLAACTAITSRNSLCSNTQQPIEIDPAFTFWTYLAVRVFIGIIGGTAFAMFEGAVIAIVRDQKADYGLQRVYGSIGGMISSPLSGLLIDYASRGKGYTDFRPAFYLYAILKVASGVLMLSIDLEFKQPAQNLLEDVITVFKNVEIVALFIACFIMGTAWGYIESFLFWLLQDLGASRSLMGITITVGGVAGLPLLVLSGPIIRKLGHANVLFIGFIFYAIRLLGYSLIYNPWLCLVFEALESVTSSLSFTAAVTYAARLSSTTTDTSVQGLLGGLYYGVGKGAGSLIGGYLMKFFGTRPTYQIFAGATLVTGCMYYLFNKFYIGKRPGSVNNDICKPQPTDVEGVKGSDKTSSPLDVATDKFENDKVLDLNHRKLVAECTDGGSDSGVENPAYNETECANGSKKDETK
Summary
Uniprot
H9J2D9
A0A2A4K094
A0A2W1BNG6
A0A194PUZ2
A0A194QYQ9
A0A212F1Z2
+ More
A0A0L7LDV0 A0A087ZWW8 A0A195B2P1 A0A158NGD7 E9J645 K7J6I3 A0A067RUB2 A0A232EM35 A0A1B6I0K6 A0A0L7R427 A0A1B6LRF6 A0A154NZV5 A0A1B6CUD4 A0A0M9A6D3 A0A151XC63 A0A026VVP7 A0A0C9PK91 E2AMH4 A0A1Y1L982 A0A1Y1L7W7 U4U650 T1I1B7 A0A195E717 A0A023EXE8 A0A0P4VUL4 A0A195F0Z5 A0A224XCK8 D6W6H6 A0A0A9W421 A0A0A9VYF4 A0A2P8YDC6 E0VXX6 N6U0Y7 E2BWD0 A0A1B6JDJ1 A0A1B6GFQ7 A0A1B6FTZ9 A0A164ZRZ8 A0A0N8DP60 A0A0P5PXH6 A0A0P5STR8 A0A0P5W8F7 A0A0P5FCU0 E9GDK2 A0A0P6A1A5 A0A0P5X3C8 A0A0N8D2I8 A0A0P6EYR9 A0A0K2UEI0 E9FWC8 A0A0P5IJR8 A0A0P5XS59 A0A0N8C4L1 A0A0P5QM10 A0A164VQC8 A0A0P5REW7 A0A0P5ZC87 A0A0P6DZ44 A0A0P5DUF4 A0A0P5JIE7 A0A0P6GGY7 A0A0P5HSR8 A0A0N8BU56 A0A164VQB3 A0A0N8AVV7 E9FWC7 A0A0P5PER1 A0A0P5K9Y1 J9KLZ0 U5EED9 A0A2R7VW56 A0A1Q3G563 A0A1Q3FZQ8
A0A0L7LDV0 A0A087ZWW8 A0A195B2P1 A0A158NGD7 E9J645 K7J6I3 A0A067RUB2 A0A232EM35 A0A1B6I0K6 A0A0L7R427 A0A1B6LRF6 A0A154NZV5 A0A1B6CUD4 A0A0M9A6D3 A0A151XC63 A0A026VVP7 A0A0C9PK91 E2AMH4 A0A1Y1L982 A0A1Y1L7W7 U4U650 T1I1B7 A0A195E717 A0A023EXE8 A0A0P4VUL4 A0A195F0Z5 A0A224XCK8 D6W6H6 A0A0A9W421 A0A0A9VYF4 A0A2P8YDC6 E0VXX6 N6U0Y7 E2BWD0 A0A1B6JDJ1 A0A1B6GFQ7 A0A1B6FTZ9 A0A164ZRZ8 A0A0N8DP60 A0A0P5PXH6 A0A0P5STR8 A0A0P5W8F7 A0A0P5FCU0 E9GDK2 A0A0P6A1A5 A0A0P5X3C8 A0A0N8D2I8 A0A0P6EYR9 A0A0K2UEI0 E9FWC8 A0A0P5IJR8 A0A0P5XS59 A0A0N8C4L1 A0A0P5QM10 A0A164VQC8 A0A0P5REW7 A0A0P5ZC87 A0A0P6DZ44 A0A0P5DUF4 A0A0P5JIE7 A0A0P6GGY7 A0A0P5HSR8 A0A0N8BU56 A0A164VQB3 A0A0N8AVV7 E9FWC7 A0A0P5PER1 A0A0P5K9Y1 J9KLZ0 U5EED9 A0A2R7VW56 A0A1Q3G563 A0A1Q3FZQ8
Pubmed
EMBL
BABH01007752
BABH01007753
NWSH01000356
PCG77072.1
KZ150007
PZC75174.1
+ More
KQ459597 KPI94955.1 KQ461150 KPJ08721.1 AGBW02010786 OWR47734.1 JTDY01001549 KOB73564.1 KQ976662 KYM78557.1 ADTU01014832 GL768221 EFZ11744.1 KK852415 KDR24400.1 NNAY01003460 OXU19372.1 GECU01027253 GECU01008791 JAS80453.1 JAS98915.1 KQ414663 KOC65506.1 GEBQ01013677 JAT26300.1 KQ434786 KZC05141.1 GEDC01020196 JAS17102.1 KQ435726 KOX78157.1 KQ982314 KYQ57900.1 KK107770 QOIP01000012 EZA47832.1 RLU16217.1 GBYB01001448 JAG71215.1 GL440813 EFN65398.1 GEZM01063958 JAV68890.1 GEZM01063957 JAV68891.1 KB631792 ERL86101.1 ACPB03016548 KQ979568 KYN20891.1 GBBI01004855 JAC13857.1 GDKW01000205 JAI56390.1 KQ981864 KYN34245.1 GFTR01007722 JAW08704.1 KQ971307 EFA11113.1 GBHO01042371 GBRD01000481 GBRD01000475 GBRD01000474 GBRD01000471 JAG01233.1 JAG65340.1 GBHO01042372 GBRD01000476 GBRD01000473 GBRD01000472 GBRD01000470 JAG01232.1 JAG65345.1 PYGN01000690 PSN42174.1 DS235842 EEB18232.1 APGK01047247 APGK01047248 KB741077 ENN74271.1 GL451103 EFN80034.1 GECU01020304 GECU01010487 JAS87402.1 JAS97219.1 GECZ01008519 JAS61250.1 GECZ01016115 JAS53654.1 LRGB01000687 KZS16680.1 GDIP01014567 JAM89148.1 GDIQ01128679 JAL23047.1 GDIP01135384 JAL68330.1 GDIP01090890 JAM12825.1 GDIQ01256918 JAJ94806.1 GL732540 EFX82094.1 GDIP01048886 JAM54829.1 GDIP01078248 JAM25467.1 GDIP01075143 JAM28572.1 GDIQ01055650 JAN39087.1 HACA01019084 CDW36445.1 GL732526 EFX88451.1 GDIQ01211548 JAK40177.1 GDIP01070044 JAM33671.1 GDIQ01115371 JAL36355.1 GDIQ01133026 JAL18700.1 LRGB01001361 KZS12548.1 GDIQ01101588 JAL50138.1 GDIP01046001 JAM57714.1 GDIQ01085197 JAN09540.1 GDIP01150572 JAJ72830.1 GDIQ01202489 JAK49236.1 GDIQ01229142 GDIQ01035027 JAN59710.1 GDIQ01223828 JAK27897.1 GDIQ01144611 GDIQ01144610 JAL07115.1 KZS12547.1 GDIQ01237803 JAK13922.1 EFX88452.1 GDIQ01129628 JAL22098.1 GDIQ01188344 JAK63381.1 ABLF02016610 ABLF02016611 ABLF02016614 GANO01004351 JAB55520.1 KK854044 PTY10165.1 GFDL01000108 JAV34937.1 GFDL01002007 JAV33038.1
KQ459597 KPI94955.1 KQ461150 KPJ08721.1 AGBW02010786 OWR47734.1 JTDY01001549 KOB73564.1 KQ976662 KYM78557.1 ADTU01014832 GL768221 EFZ11744.1 KK852415 KDR24400.1 NNAY01003460 OXU19372.1 GECU01027253 GECU01008791 JAS80453.1 JAS98915.1 KQ414663 KOC65506.1 GEBQ01013677 JAT26300.1 KQ434786 KZC05141.1 GEDC01020196 JAS17102.1 KQ435726 KOX78157.1 KQ982314 KYQ57900.1 KK107770 QOIP01000012 EZA47832.1 RLU16217.1 GBYB01001448 JAG71215.1 GL440813 EFN65398.1 GEZM01063958 JAV68890.1 GEZM01063957 JAV68891.1 KB631792 ERL86101.1 ACPB03016548 KQ979568 KYN20891.1 GBBI01004855 JAC13857.1 GDKW01000205 JAI56390.1 KQ981864 KYN34245.1 GFTR01007722 JAW08704.1 KQ971307 EFA11113.1 GBHO01042371 GBRD01000481 GBRD01000475 GBRD01000474 GBRD01000471 JAG01233.1 JAG65340.1 GBHO01042372 GBRD01000476 GBRD01000473 GBRD01000472 GBRD01000470 JAG01232.1 JAG65345.1 PYGN01000690 PSN42174.1 DS235842 EEB18232.1 APGK01047247 APGK01047248 KB741077 ENN74271.1 GL451103 EFN80034.1 GECU01020304 GECU01010487 JAS87402.1 JAS97219.1 GECZ01008519 JAS61250.1 GECZ01016115 JAS53654.1 LRGB01000687 KZS16680.1 GDIP01014567 JAM89148.1 GDIQ01128679 JAL23047.1 GDIP01135384 JAL68330.1 GDIP01090890 JAM12825.1 GDIQ01256918 JAJ94806.1 GL732540 EFX82094.1 GDIP01048886 JAM54829.1 GDIP01078248 JAM25467.1 GDIP01075143 JAM28572.1 GDIQ01055650 JAN39087.1 HACA01019084 CDW36445.1 GL732526 EFX88451.1 GDIQ01211548 JAK40177.1 GDIP01070044 JAM33671.1 GDIQ01115371 JAL36355.1 GDIQ01133026 JAL18700.1 LRGB01001361 KZS12548.1 GDIQ01101588 JAL50138.1 GDIP01046001 JAM57714.1 GDIQ01085197 JAN09540.1 GDIP01150572 JAJ72830.1 GDIQ01202489 JAK49236.1 GDIQ01229142 GDIQ01035027 JAN59710.1 GDIQ01223828 JAK27897.1 GDIQ01144611 GDIQ01144610 JAL07115.1 KZS12547.1 GDIQ01237803 JAK13922.1 EFX88452.1 GDIQ01129628 JAL22098.1 GDIQ01188344 JAK63381.1 ABLF02016610 ABLF02016611 ABLF02016614 GANO01004351 JAB55520.1 KK854044 PTY10165.1 GFDL01000108 JAV34937.1 GFDL01002007 JAV33038.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000005203 UP000078540 UP000005205 UP000002358 UP000027135 UP000215335 UP000053825 UP000076502 UP000053105 UP000075809 UP000053097 UP000279307 UP000000311 UP000030742 UP000015103 UP000078492 UP000078541 UP000007266 UP000245037 UP000009046 UP000019118 UP000008237 UP000076858 UP000000305 UP000007819
UP000005203 UP000078540 UP000005205 UP000002358 UP000027135 UP000215335 UP000053825 UP000076502 UP000053105 UP000075809 UP000053097 UP000279307 UP000000311 UP000030742 UP000015103 UP000078492 UP000078541 UP000007266 UP000245037 UP000009046 UP000019118 UP000008237 UP000076858 UP000000305 UP000007819
PRIDE
Pfam
PF12832 MFS_1_like
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9J2D9
A0A2A4K094
A0A2W1BNG6
A0A194PUZ2
A0A194QYQ9
A0A212F1Z2
+ More
A0A0L7LDV0 A0A087ZWW8 A0A195B2P1 A0A158NGD7 E9J645 K7J6I3 A0A067RUB2 A0A232EM35 A0A1B6I0K6 A0A0L7R427 A0A1B6LRF6 A0A154NZV5 A0A1B6CUD4 A0A0M9A6D3 A0A151XC63 A0A026VVP7 A0A0C9PK91 E2AMH4 A0A1Y1L982 A0A1Y1L7W7 U4U650 T1I1B7 A0A195E717 A0A023EXE8 A0A0P4VUL4 A0A195F0Z5 A0A224XCK8 D6W6H6 A0A0A9W421 A0A0A9VYF4 A0A2P8YDC6 E0VXX6 N6U0Y7 E2BWD0 A0A1B6JDJ1 A0A1B6GFQ7 A0A1B6FTZ9 A0A164ZRZ8 A0A0N8DP60 A0A0P5PXH6 A0A0P5STR8 A0A0P5W8F7 A0A0P5FCU0 E9GDK2 A0A0P6A1A5 A0A0P5X3C8 A0A0N8D2I8 A0A0P6EYR9 A0A0K2UEI0 E9FWC8 A0A0P5IJR8 A0A0P5XS59 A0A0N8C4L1 A0A0P5QM10 A0A164VQC8 A0A0P5REW7 A0A0P5ZC87 A0A0P6DZ44 A0A0P5DUF4 A0A0P5JIE7 A0A0P6GGY7 A0A0P5HSR8 A0A0N8BU56 A0A164VQB3 A0A0N8AVV7 E9FWC7 A0A0P5PER1 A0A0P5K9Y1 J9KLZ0 U5EED9 A0A2R7VW56 A0A1Q3G563 A0A1Q3FZQ8
A0A0L7LDV0 A0A087ZWW8 A0A195B2P1 A0A158NGD7 E9J645 K7J6I3 A0A067RUB2 A0A232EM35 A0A1B6I0K6 A0A0L7R427 A0A1B6LRF6 A0A154NZV5 A0A1B6CUD4 A0A0M9A6D3 A0A151XC63 A0A026VVP7 A0A0C9PK91 E2AMH4 A0A1Y1L982 A0A1Y1L7W7 U4U650 T1I1B7 A0A195E717 A0A023EXE8 A0A0P4VUL4 A0A195F0Z5 A0A224XCK8 D6W6H6 A0A0A9W421 A0A0A9VYF4 A0A2P8YDC6 E0VXX6 N6U0Y7 E2BWD0 A0A1B6JDJ1 A0A1B6GFQ7 A0A1B6FTZ9 A0A164ZRZ8 A0A0N8DP60 A0A0P5PXH6 A0A0P5STR8 A0A0P5W8F7 A0A0P5FCU0 E9GDK2 A0A0P6A1A5 A0A0P5X3C8 A0A0N8D2I8 A0A0P6EYR9 A0A0K2UEI0 E9FWC8 A0A0P5IJR8 A0A0P5XS59 A0A0N8C4L1 A0A0P5QM10 A0A164VQC8 A0A0P5REW7 A0A0P5ZC87 A0A0P6DZ44 A0A0P5DUF4 A0A0P5JIE7 A0A0P6GGY7 A0A0P5HSR8 A0A0N8BU56 A0A164VQB3 A0A0N8AVV7 E9FWC7 A0A0P5PER1 A0A0P5K9Y1 J9KLZ0 U5EED9 A0A2R7VW56 A0A1Q3G563 A0A1Q3FZQ8
Ontologies
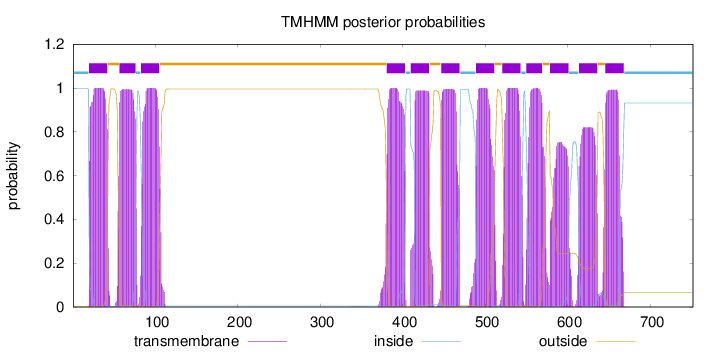
Topology
Length:
752
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
250.87625
Exp number, first 60 AAs:
27.44983
Total prob of N-in:
0.99882
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 56
TMhelix
57 - 76
inside
77 - 82
TMhelix
83 - 105
outside
106 - 380
TMhelix
381 - 403
inside
404 - 409
TMhelix
410 - 432
outside
433 - 446
TMhelix
447 - 469
inside
470 - 488
TMhelix
489 - 511
outside
512 - 520
TMhelix
521 - 543
inside
544 - 549
TMhelix
550 - 569
outside
570 - 578
TMhelix
579 - 601
inside
602 - 613
TMhelix
614 - 636
outside
637 - 645
TMhelix
646 - 668
inside
669 - 752
Population Genetic Test Statistics
Pi
204.306563
Theta
174.115521
Tajima's D
0.863501
CLR
0.070117
CSRT
0.617469126543673
Interpretation
Uncertain
