Gene
KWMTBOMO02825
Pre Gene Modal
BGIBMGA003679
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101737507_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.19
Sequence
CDS
ATGCACGCACGGCTCCGACCCGCCTGCCTGCCGCCGGCGTACCACATGCCGCCCCCCGGACGACACTGCACGGTCGTCGGCTGGGGGCAATTGTACGAGCACGAACGAGTTTTCCCGGACACCCTGCAAGAGGTGGAGTTACCGGTGATATCGAGCGCGGAGTGCCGGCGTCGCACGCGCTTGCTCCCCTTGTACAGAGTCACCGACGACATGTTCTGCGCCGGATACGACCGCGGCGGCCGCGACGCTTGTCTCGGGGATTCGGGGGGACCACTAATGTGTCAGGAATCCGGAAGATGGTATATCTACGGTGTGACCAGCAACGGATACGGTTGCGCTCGGGCGAATCGCCCGGGCGTCTACACCAAAGTCTCAAACTACATCGACTGGATCGACTCGGTGATCGCTACCAACACTCCAGCGCACAACGACACCGAACTCGACGAGAACGAATCGAACGAGGAGTTCTACGCGGACTTAGAAGTCGCAGAGAACCGGCGACATAGAGTAGACACTTGCAGAGGGCACAGGTGTCCCCTCGGCGAGTGCCTGCCGCCGTCGAGCGTGTGCAACGGGTTTCTAGAATGTTCTGACGGCAGCGACGAATGGCAATGCAGCAAGCCCCTAAACAACACTAGCTTAGATCCCGATTAA
Protein
MHARLRPACLPPAYHMPPPGRHCTVVGWGQLYEHERVFPDTLQEVELPVISSAECRRRTRLLPLYRVTDDMFCAGYDRGGRDACLGDSGGPLMCQESGRWYIYGVTSNGYGCARANRPGVYTKVSNYIDWIDSVIATNTPAHNDTELDENESNEEFYADLEVAENRRHRVDTCRGHRCPLGECLPPSSVCNGFLECSDGSDEWQCSKPLNNTSLDPD
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9J2E2
A0A2H1VXR2
A0A2W1BPE2
A0A2A4IU68
A0A194PNE1
A0A212F1W5
+ More
A0A194QTT5 A0A2P8XPL7 U4U826 A0A1V1FIS7 A0A1B6I646 A0A139WNH6 D6W6H7 A0A1B6E874 N6U6S4 A0A023EYC6 A0A0A9XL31 A0A067RUA7 T1H7X5 A0A1Y1KM35 A0A0P4W7C6 A0A0P5ENW6 A0A0P6JIW2 A0A0N8CH09 A0A0P5J903 A0A0P6CVY3 A0A0P5ACF8 A0A0P5KC84 E0VXL2 A0A2R7W410 A0A151IQL6 A0A0M9A8L1 A0A154NZY0 A0A1D2N774 A0A195E7U2 A0A158NFX9 A0A195B2P7 A0A151XC08 A0A0C9RZE6 A0A0L7R3U7 F4X6W9 A0A195F250 E2BWC8 E9H2M8 A0A3L8D802 A0A026VY88 A0A087T3Z0 A0A232F4T1 T1ITP1 K7JBQ3 A0A1Y3B4S3 A0A023F157
A0A194QTT5 A0A2P8XPL7 U4U826 A0A1V1FIS7 A0A1B6I646 A0A139WNH6 D6W6H7 A0A1B6E874 N6U6S4 A0A023EYC6 A0A0A9XL31 A0A067RUA7 T1H7X5 A0A1Y1KM35 A0A0P4W7C6 A0A0P5ENW6 A0A0P6JIW2 A0A0N8CH09 A0A0P5J903 A0A0P6CVY3 A0A0P5ACF8 A0A0P5KC84 E0VXL2 A0A2R7W410 A0A151IQL6 A0A0M9A8L1 A0A154NZY0 A0A1D2N774 A0A195E7U2 A0A158NFX9 A0A195B2P7 A0A151XC08 A0A0C9RZE6 A0A0L7R3U7 F4X6W9 A0A195F250 E2BWC8 E9H2M8 A0A3L8D802 A0A026VY88 A0A087T3Z0 A0A232F4T1 T1ITP1 K7JBQ3 A0A1Y3B4S3 A0A023F157
Pubmed
EMBL
BABH01007759
ODYU01005033
SOQ45516.1
KZ150007
PZC75177.1
NWSH01006316
+ More
PCG63517.1 KQ459597 KPI94951.1 AGBW02010786 OWR47730.1 KQ461150 KPJ08724.1 PYGN01001574 PSN33951.1 KB631792 ERL86100.1 FX985311 BAX07324.1 GECU01025329 JAS82377.1 KQ971307 KYB29568.1 EFA11570.2 GEDC01003175 JAS34123.1 APGK01047249 APGK01047250 KB741077 ENN74272.1 GBBI01004620 JAC14092.1 GBHO01024096 JAG19508.1 KK852415 KDR24395.1 ACPB03016548 ACPB03016549 GEZM01085901 JAV59907.1 GDRN01085776 JAI61287.1 GDIP01145317 JAJ78085.1 GDIQ01006518 JAN88219.1 GDIP01132575 JAL71139.1 GDIQ01203585 JAK48140.1 GDIP01008475 JAM95240.1 GDIP01201862 GDIP01108923 LRGB01000311 JAJ21540.1 KZS19905.1 GDIQ01186345 JAK65380.1 DS235833 EEB18118.1 KK854305 PTY14412.1 KQ976780 KYN08333.1 KQ435726 KOX78153.1 KQ434786 KZC05142.1 LJIJ01000172 ODN01091.1 KQ979568 KYN20889.1 ADTU01014831 ADTU01014832 KQ976662 KYM78555.1 KQ982314 KYQ57902.1 GBYB01014785 JAG84552.1 KQ414663 KOC65504.1 GL888818 EGI57901.1 KQ981864 KYN34247.1 GL451103 EFN80032.1 GL732586 EFX74036.1 QOIP01000012 RLU16222.1 KK107770 EZA47834.1 KK113290 KFM59829.1 NNAY01001018 OXU25473.1 JH431493 AAZX01001290 MUJZ01044445 OTF74958.1 GBBI01003956 JAC14756.1
PCG63517.1 KQ459597 KPI94951.1 AGBW02010786 OWR47730.1 KQ461150 KPJ08724.1 PYGN01001574 PSN33951.1 KB631792 ERL86100.1 FX985311 BAX07324.1 GECU01025329 JAS82377.1 KQ971307 KYB29568.1 EFA11570.2 GEDC01003175 JAS34123.1 APGK01047249 APGK01047250 KB741077 ENN74272.1 GBBI01004620 JAC14092.1 GBHO01024096 JAG19508.1 KK852415 KDR24395.1 ACPB03016548 ACPB03016549 GEZM01085901 JAV59907.1 GDRN01085776 JAI61287.1 GDIP01145317 JAJ78085.1 GDIQ01006518 JAN88219.1 GDIP01132575 JAL71139.1 GDIQ01203585 JAK48140.1 GDIP01008475 JAM95240.1 GDIP01201862 GDIP01108923 LRGB01000311 JAJ21540.1 KZS19905.1 GDIQ01186345 JAK65380.1 DS235833 EEB18118.1 KK854305 PTY14412.1 KQ976780 KYN08333.1 KQ435726 KOX78153.1 KQ434786 KZC05142.1 LJIJ01000172 ODN01091.1 KQ979568 KYN20889.1 ADTU01014831 ADTU01014832 KQ976662 KYM78555.1 KQ982314 KYQ57902.1 GBYB01014785 JAG84552.1 KQ414663 KOC65504.1 GL888818 EGI57901.1 KQ981864 KYN34247.1 GL451103 EFN80032.1 GL732586 EFX74036.1 QOIP01000012 RLU16222.1 KK107770 EZA47834.1 KK113290 KFM59829.1 NNAY01001018 OXU25473.1 JH431493 AAZX01001290 MUJZ01044445 OTF74958.1 GBBI01003956 JAC14756.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000245037
+ More
UP000030742 UP000007266 UP000019118 UP000027135 UP000015103 UP000076858 UP000009046 UP000078542 UP000053105 UP000076502 UP000094527 UP000078492 UP000005205 UP000078540 UP000075809 UP000053825 UP000007755 UP000078541 UP000008237 UP000000305 UP000279307 UP000053097 UP000054359 UP000215335 UP000002358
UP000030742 UP000007266 UP000019118 UP000027135 UP000015103 UP000076858 UP000009046 UP000078542 UP000053105 UP000076502 UP000094527 UP000078492 UP000005205 UP000078540 UP000075809 UP000053825 UP000007755 UP000078541 UP000008237 UP000000305 UP000279307 UP000053097 UP000054359 UP000215335 UP000002358
PRIDE
Interpro
IPR036055
LDL_receptor-like_sf
+ More
IPR002172 LDrepeatLR_classA_rpt
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR001190 SRCR
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR000082 SEA_dom
IPR033116 TRYPSIN_SER
IPR023415 LDLR_class-A_CS
IPR036772 SRCR-like_dom_sf
IPR036364 SEA_dom_sf
IPR036277 SMC_hinge_sf
IPR038892 SMCHD1
IPR010935 SMC_hinge
IPR036890 HATPase_C_sf
IPR003594 HATPase_C
IPR015420 Peptidase_S1A_nudel
IPR002172 LDrepeatLR_classA_rpt
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR001190 SRCR
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR000082 SEA_dom
IPR033116 TRYPSIN_SER
IPR023415 LDLR_class-A_CS
IPR036772 SRCR-like_dom_sf
IPR036364 SEA_dom_sf
IPR036277 SMC_hinge_sf
IPR038892 SMCHD1
IPR010935 SMC_hinge
IPR036890 HATPase_C_sf
IPR003594 HATPase_C
IPR015420 Peptidase_S1A_nudel
SUPFAM
Gene 3D
ProteinModelPortal
H9J2E2
A0A2H1VXR2
A0A2W1BPE2
A0A2A4IU68
A0A194PNE1
A0A212F1W5
+ More
A0A194QTT5 A0A2P8XPL7 U4U826 A0A1V1FIS7 A0A1B6I646 A0A139WNH6 D6W6H7 A0A1B6E874 N6U6S4 A0A023EYC6 A0A0A9XL31 A0A067RUA7 T1H7X5 A0A1Y1KM35 A0A0P4W7C6 A0A0P5ENW6 A0A0P6JIW2 A0A0N8CH09 A0A0P5J903 A0A0P6CVY3 A0A0P5ACF8 A0A0P5KC84 E0VXL2 A0A2R7W410 A0A151IQL6 A0A0M9A8L1 A0A154NZY0 A0A1D2N774 A0A195E7U2 A0A158NFX9 A0A195B2P7 A0A151XC08 A0A0C9RZE6 A0A0L7R3U7 F4X6W9 A0A195F250 E2BWC8 E9H2M8 A0A3L8D802 A0A026VY88 A0A087T3Z0 A0A232F4T1 T1ITP1 K7JBQ3 A0A1Y3B4S3 A0A023F157
A0A194QTT5 A0A2P8XPL7 U4U826 A0A1V1FIS7 A0A1B6I646 A0A139WNH6 D6W6H7 A0A1B6E874 N6U6S4 A0A023EYC6 A0A0A9XL31 A0A067RUA7 T1H7X5 A0A1Y1KM35 A0A0P4W7C6 A0A0P5ENW6 A0A0P6JIW2 A0A0N8CH09 A0A0P5J903 A0A0P6CVY3 A0A0P5ACF8 A0A0P5KC84 E0VXL2 A0A2R7W410 A0A151IQL6 A0A0M9A8L1 A0A154NZY0 A0A1D2N774 A0A195E7U2 A0A158NFX9 A0A195B2P7 A0A151XC08 A0A0C9RZE6 A0A0L7R3U7 F4X6W9 A0A195F250 E2BWC8 E9H2M8 A0A3L8D802 A0A026VY88 A0A087T3Z0 A0A232F4T1 T1ITP1 K7JBQ3 A0A1Y3B4S3 A0A023F157
PDB
4DGJ
E-value=1.01836e-33,
Score=355
Ontologies
GO
PANTHER
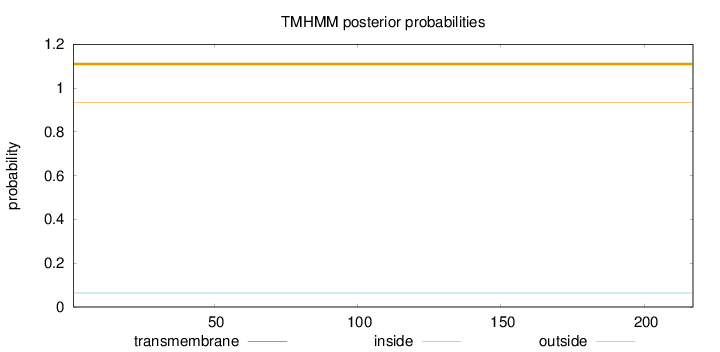
Topology
Length:
217
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00074
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06448
outside
1 - 217
Population Genetic Test Statistics
Pi
259.245064
Theta
193.315404
Tajima's D
1.443496
CLR
0.192995
CSRT
0.775461226938653
Interpretation
Uncertain
