Gene
KWMTBOMO02824 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003515
Annotation
PREDICTED:_adenylyl_cyclase-associated_protein_1_isoform_X1_[Bombyx_mori]
Full name
Adenylyl cyclase-associated protein
Location in the cell
Nuclear Reliability : 2.153
Sequence
CDS
ATGGAAGAAAGTGCACAAAGAAACCTAATGAACTTAATTAAAATGTTCGAAGGACAATCAAATATTCAAAACGAAGGAGGAGATAGTGTTGAGTCAACACACGAAGCTGTTAATACGGTGATGGGTAACGAAGTCGAGGGCACCGCAGCTGCTCACGGCTCGGCATCGGATCTCGAGACGCTGATCGAGCGTTTGGAACGCGTTACCTCGCGATTAGAACGTCTGCCGCTGCTGCGAGCTCATACTCCTGCAGCTCCTCATTCGCCGGCGCCTTCTCCAGTCTCGCTGAACACACCACCGCCCCAAGAATCGCTCGAACCAATACCATCGGACAACATGAGTGTCAACGGATATCAAGACCTTGTCCAGGGTCCCCTCCGCGCCTATCTCCAACTGTCGCAGCAGATTGGCGGAGACGTTGCTACTCAAGCTAACCTCGTGAATGCTGCTTTTCAAGCGCAATCCCGCTACATACAACTGGCGGCAACAAAAGCCAAGCCTTCCCAAGCTGAGGAAATGCAGCTTCTGACTCCGACCAGCGAACAGATATCTGCTATCCAGCAATATCGTGAAAAGAATCGCGCCTCGCAGTTCTTTAATCATCTCTCGGCGATATCCGAGAGTATCCCGGCTTTGGGATGGGTAGCCGTTGCTCCCGCCCCGGCTCCGTACGTTAAGGAAATGAATGACGCCGGCCAGTTCTACACTAACCGCGTGCTCAAGGATTGGAAGGAGAAAGACAAGACCCACGTAGAGTGGTGCCGCGCTTGGGTCCAACTGCTGTCCGATTTGCAAGCGTACGTCAAACAGTTTCACACAACAGGACTTGTGTGGTCTGGCAAGGGCAGTGCTGGTGCCCCCCCACCGCCTCCCCCGGGAGGACTTCCCCCTCCGCCGCCCGTGCTGCCAGACGTGGACTTCTCCAACTTGTCGCTCGATGACCGCAGCGCTCTCTTCGCCGAAATCAATCGAGGCGAAGCCATAACTAGCTCCTTGCGCAAAGTGACGCCGGACATGCAGACCCACAAGAATCCGACTCTCCGTCAAGGCCCGGCGCCGTTCAAAGCGACGTCGGCCGCCGGACACAAGCCGCTGCCGCAGCCCGGCGCTGGACAACTGGACAAGCCGCCCGTCTTCACTCGGGACGGCAAGAAGTGGCTTATCGAGTACCAAAAGGGCAACTCCAATTTAGTGGTCGAGAACGCGGACATGAACAACGTTGTGTACATGTTCCGTTGTCGCGACTCCGCCCTGACGGTCCGCGGGAAGGTCAACGGGGTCGTGCTGGACTCGTGCACCAAGTGCGCGGTCGTGTTCGACAACCTCGTATCCAGCATCGAGTTCGTCAACTGCCAGTCCGTACAGATGCAGGTATTGGGCAAAGTTCCAACCGTTTCCATCGACAAGACCGACGGATGTCAGATATATCTGTCGCCGGAATCGCTGGACGTCGAAATCGTCAGCTCCAAGTCATCGGAGATGAACGTTTTGATACCCAAGGCTAATGGCGATTACACCGAACACGCGATCCCCGAGCAGTTCAAGACGGTACTGAACAAGCCGGCCGGCCTCACCACGACGCCCGTCGAGAACAAGGGCTAA
Protein
MEESAQRNLMNLIKMFEGQSNIQNEGGDSVESTHEAVNTVMGNEVEGTAAAHGSASDLETLIERLERVTSRLERLPLLRAHTPAAPHSPAPSPVSLNTPPPQESLEPIPSDNMSVNGYQDLVQGPLRAYLQLSQQIGGDVATQANLVNAAFQAQSRYIQLAATKAKPSQAEEMQLLTPTSEQISAIQQYREKNRASQFFNHLSAISESIPALGWVAVAPAPAPYVKEMNDAGQFYTNRVLKDWKEKDKTHVEWCRAWVQLLSDLQAYVKQFHTTGLVWSGKGSAGAPPPPPPGGLPPPPPVLPDVDFSNLSLDDRSALFAEINRGEAITSSLRKVTPDMQTHKNPTLRQGPAPFKATSAAGHKPLPQPGAGQLDKPPVFTRDGKKWLIEYQKGNSNLVVENADMNNVVYMFRCRDSALTVRGKVNGVVLDSCTKCAVVFDNLVSSIEFVNCQSVQMQVLGKVPTVSIDKTDGCQIYLSPESLDVEIVSSKSSEMNVLIPKANGDYTEHAIPEQFKTVLNKPAGLTTTPVENKG
Summary
Similarity
Belongs to the CAP family.
Feature
chain Adenylyl cyclase-associated protein
Uniprot
A0A2A4IVW7
A0A194QUR9
A0A2W1BJX1
A0A212F1V6
A0A1E1WD75
A0A194PPP8
+ More
D6W6L7 A0A154P1C4 A0A1L8E5T3 A0A1L8E5U4 A0A1L8E573 E2ARB8 A0A1B6DH10 A0A0M9A6C6 A0A1B6C4R3 E2BWC7 A0A151XC21 A0A1B6IJ81 A0A1B6FWV4 A0A151IQD3 A0A336KDK4 A0A158NGD4 B4KEU8 A0A087ZPC0 A0A1I8NBN4 A0A195F1A6 A0A195E7R5 T1PCY9 A0A336KDG3 W8BFG0 T1PLK0 A0A1B6F8A1 A0A3L8D7K4 Q16K93 A0A1B6JNG9 A0A026VVZ0 A0A1Q3FG29 A0A0L7R3P1 A0A1Q3FG63 W8BT17 A0A0Q9XIS5 A0A1I8PFE4 U5EUZ2 A0A1I8PF50 A0A023EVL8 A0A034VE93 A0A0T6B7Q4 A0A0P6K160 A0A0A9W4T0 A0A034VGY7 A0A1A9UTX7 A0A034VGB4 A0A1B0FML2 A0A1A9Z5Y4 A0A1Y1LPT5 A0A1W4X8E2 V5I8H4 A0A224XIV7 A0A2M4A5S5 A0A0P4VL56 J3JUS4 A0A2M4A5M6 A0A2M4A5M3 A0A2M4A5W1 A0A2M4A5I8 A0A0C9Q702 A0A0V0G8D7 A0A1Q3FFZ7 A0A1Q3FGF1 A0A1Q3FGH8 A0A0K8VLI6 A0A1L8EBC2 A0A0K8UJZ0 A0A1I8PFE1 A0A0R1DII5 A0A1L8EBF1 A0A1L8EBC1 A0A034VGZ2 A0A1Q3FG80 A0A1L8EBS0 A0A1Q3FFY7 A0A182MQE0 A0A1B0BDT3 A0A1A9XYS8 A0A0P8ZET0 A0A023F3J3 A0A0L0CFZ8 A0A1Q3FG11 A0A0R1DIJ5 A0A1Q3FFU7 A0A1A9WUC6 Q7KN55 Q9VPX7 A0A069DT58 A0A0M4E8Z6 A0A0Q9WCU9 A0A0Q9WNI2 A0A1S4H405 B4JDF4
D6W6L7 A0A154P1C4 A0A1L8E5T3 A0A1L8E5U4 A0A1L8E573 E2ARB8 A0A1B6DH10 A0A0M9A6C6 A0A1B6C4R3 E2BWC7 A0A151XC21 A0A1B6IJ81 A0A1B6FWV4 A0A151IQD3 A0A336KDK4 A0A158NGD4 B4KEU8 A0A087ZPC0 A0A1I8NBN4 A0A195F1A6 A0A195E7R5 T1PCY9 A0A336KDG3 W8BFG0 T1PLK0 A0A1B6F8A1 A0A3L8D7K4 Q16K93 A0A1B6JNG9 A0A026VVZ0 A0A1Q3FG29 A0A0L7R3P1 A0A1Q3FG63 W8BT17 A0A0Q9XIS5 A0A1I8PFE4 U5EUZ2 A0A1I8PF50 A0A023EVL8 A0A034VE93 A0A0T6B7Q4 A0A0P6K160 A0A0A9W4T0 A0A034VGY7 A0A1A9UTX7 A0A034VGB4 A0A1B0FML2 A0A1A9Z5Y4 A0A1Y1LPT5 A0A1W4X8E2 V5I8H4 A0A224XIV7 A0A2M4A5S5 A0A0P4VL56 J3JUS4 A0A2M4A5M6 A0A2M4A5M3 A0A2M4A5W1 A0A2M4A5I8 A0A0C9Q702 A0A0V0G8D7 A0A1Q3FFZ7 A0A1Q3FGF1 A0A1Q3FGH8 A0A0K8VLI6 A0A1L8EBC2 A0A0K8UJZ0 A0A1I8PFE1 A0A0R1DII5 A0A1L8EBF1 A0A1L8EBC1 A0A034VGZ2 A0A1Q3FG80 A0A1L8EBS0 A0A1Q3FFY7 A0A182MQE0 A0A1B0BDT3 A0A1A9XYS8 A0A0P8ZET0 A0A023F3J3 A0A0L0CFZ8 A0A1Q3FG11 A0A0R1DIJ5 A0A1Q3FFU7 A0A1A9WUC6 Q7KN55 Q9VPX7 A0A069DT58 A0A0M4E8Z6 A0A0Q9WCU9 A0A0Q9WNI2 A0A1S4H405 B4JDF4
Pubmed
26354079
28756777
22118469
18362917
19820115
20798317
+ More
21347285 17994087 25315136 24495485 30249741 17510324 24508170 24945155 25348373 26999592 25401762 26823975 28004739 27129103 22516182 23537049 17550304 25474469 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 12364791
21347285 17994087 25315136 24495485 30249741 17510324 24508170 24945155 25348373 26999592 25401762 26823975 28004739 27129103 22516182 23537049 17550304 25474469 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 12364791
EMBL
NWSH01006316
PCG63518.1
KQ461150
KPJ08725.1
KZ150007
PZC75178.1
+ More
AGBW02010786 OWR47728.1 GDQN01006149 GDQN01000551 JAT84905.1 JAT90503.1 KQ459597 KPI94948.1 KQ971307 EFA10858.2 KQ434786 KZC05144.1 GFDF01000212 JAV13872.1 GFDF01000202 JAV13882.1 GFDF01000203 JAV13881.1 GL442017 EFN64022.1 GEDC01012322 GEDC01010354 GEDC01003737 JAS24976.1 JAS26944.1 JAS33561.1 KQ435726 KOX78150.1 GEDC01028924 JAS08374.1 GL451103 EFN80031.1 KQ982314 KYQ57903.1 GECU01029575 GECU01020724 JAS78131.1 JAS86982.1 GECZ01015081 JAS54688.1 KQ976780 KYN08330.1 UFQS01000295 UFQT01000295 SSX02569.1 SSX22943.1 ADTU01014830 ADTU01014831 CH933807 EDW12998.1 KQ981864 KYN34248.1 KQ979568 KYN20887.1 KA646657 AFP61286.1 SSX02571.1 SSX22945.1 GAMC01006505 JAC00051.1 KA649589 AFP64218.1 GECZ01023379 JAS46390.1 QOIP01000012 RLU16221.1 CH477971 EAT34722.1 GECU01006987 JAT00720.1 KK107770 EZA47835.1 GFDL01008535 JAV26510.1 KQ414663 KOC65502.1 GFDL01008499 JAV26546.1 GAMC01006502 JAC00054.1 KRG03539.1 GANO01002111 JAB57760.1 GAPW01000512 JAC13086.1 GAKP01018143 JAC40809.1 LJIG01009284 KRT83388.1 GDUN01000041 JAN95878.1 GBHO01042127 GBHO01042124 GBHO01042121 GBRD01012844 GBRD01012842 GBRD01012841 GDHC01004661 GDHC01002555 JAG01477.1 JAG01480.1 JAG01483.1 JAG52982.1 JAQ13968.1 JAQ16074.1 GAKP01018144 JAC40808.1 GAKP01018142 GAKP01018141 GAKP01018140 JAC40810.1 CCAG010018213 CCAG010018214 GEZM01050241 JAV75659.1 GALX01004680 JAB63786.1 GFTR01006678 JAW09748.1 GGFK01002808 MBW36129.1 GDKW01003292 JAI53303.1 APGK01041327 BT126989 KB740993 KB632256 AEE61951.1 ENN76015.1 ERL90760.1 GGFK01002734 MBW36055.1 GGFK01002786 MBW36107.1 GGFK01002820 MBW36141.1 GGFK01002732 MBW36053.1 GBYB01009913 JAG79680.1 GECL01001772 JAP04352.1 GFDL01008571 JAV26474.1 GFDL01008424 JAV26621.1 GFDL01008443 JAV26602.1 GDHF01012573 JAI39741.1 GFDG01002742 JAV16057.1 GDHF01025654 JAI26660.1 CM000157 KRJ97060.1 GFDG01002741 JAV16058.1 GFDG01002739 JAV16060.1 GAKP01018139 JAC40813.1 GFDL01008485 JAV26560.1 GFDG01002740 JAV16059.1 GFDL01008590 JAV26455.1 AXCM01000013 JXJN01012663 JXJN01012664 JXJN01012665 JXJN01012666 JXJN01012667 JXJN01012668 CH902620 KPU73197.1 GBBI01002816 JAC15896.1 JRES01000438 KNC31171.1 GFDL01008558 JAV26487.1 KRJ97061.1 GFDL01008621 JAV26424.1 AF132566 AAD27865.2 AE014134 AAF51408.2 GBGD01001611 JAC87278.1 CP012523 ALC38576.1 CH940649 KRF82006.1 KRF82005.1 AAAB01008980 CH916368 EDW03324.1
AGBW02010786 OWR47728.1 GDQN01006149 GDQN01000551 JAT84905.1 JAT90503.1 KQ459597 KPI94948.1 KQ971307 EFA10858.2 KQ434786 KZC05144.1 GFDF01000212 JAV13872.1 GFDF01000202 JAV13882.1 GFDF01000203 JAV13881.1 GL442017 EFN64022.1 GEDC01012322 GEDC01010354 GEDC01003737 JAS24976.1 JAS26944.1 JAS33561.1 KQ435726 KOX78150.1 GEDC01028924 JAS08374.1 GL451103 EFN80031.1 KQ982314 KYQ57903.1 GECU01029575 GECU01020724 JAS78131.1 JAS86982.1 GECZ01015081 JAS54688.1 KQ976780 KYN08330.1 UFQS01000295 UFQT01000295 SSX02569.1 SSX22943.1 ADTU01014830 ADTU01014831 CH933807 EDW12998.1 KQ981864 KYN34248.1 KQ979568 KYN20887.1 KA646657 AFP61286.1 SSX02571.1 SSX22945.1 GAMC01006505 JAC00051.1 KA649589 AFP64218.1 GECZ01023379 JAS46390.1 QOIP01000012 RLU16221.1 CH477971 EAT34722.1 GECU01006987 JAT00720.1 KK107770 EZA47835.1 GFDL01008535 JAV26510.1 KQ414663 KOC65502.1 GFDL01008499 JAV26546.1 GAMC01006502 JAC00054.1 KRG03539.1 GANO01002111 JAB57760.1 GAPW01000512 JAC13086.1 GAKP01018143 JAC40809.1 LJIG01009284 KRT83388.1 GDUN01000041 JAN95878.1 GBHO01042127 GBHO01042124 GBHO01042121 GBRD01012844 GBRD01012842 GBRD01012841 GDHC01004661 GDHC01002555 JAG01477.1 JAG01480.1 JAG01483.1 JAG52982.1 JAQ13968.1 JAQ16074.1 GAKP01018144 JAC40808.1 GAKP01018142 GAKP01018141 GAKP01018140 JAC40810.1 CCAG010018213 CCAG010018214 GEZM01050241 JAV75659.1 GALX01004680 JAB63786.1 GFTR01006678 JAW09748.1 GGFK01002808 MBW36129.1 GDKW01003292 JAI53303.1 APGK01041327 BT126989 KB740993 KB632256 AEE61951.1 ENN76015.1 ERL90760.1 GGFK01002734 MBW36055.1 GGFK01002786 MBW36107.1 GGFK01002820 MBW36141.1 GGFK01002732 MBW36053.1 GBYB01009913 JAG79680.1 GECL01001772 JAP04352.1 GFDL01008571 JAV26474.1 GFDL01008424 JAV26621.1 GFDL01008443 JAV26602.1 GDHF01012573 JAI39741.1 GFDG01002742 JAV16057.1 GDHF01025654 JAI26660.1 CM000157 KRJ97060.1 GFDG01002741 JAV16058.1 GFDG01002739 JAV16060.1 GAKP01018139 JAC40813.1 GFDL01008485 JAV26560.1 GFDG01002740 JAV16059.1 GFDL01008590 JAV26455.1 AXCM01000013 JXJN01012663 JXJN01012664 JXJN01012665 JXJN01012666 JXJN01012667 JXJN01012668 CH902620 KPU73197.1 GBBI01002816 JAC15896.1 JRES01000438 KNC31171.1 GFDL01008558 JAV26487.1 KRJ97061.1 GFDL01008621 JAV26424.1 AF132566 AAD27865.2 AE014134 AAF51408.2 GBGD01001611 JAC87278.1 CP012523 ALC38576.1 CH940649 KRF82006.1 KRF82005.1 AAAB01008980 CH916368 EDW03324.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000007266
UP000076502
+ More
UP000000311 UP000053105 UP000008237 UP000075809 UP000078542 UP000005205 UP000009192 UP000005203 UP000095301 UP000078541 UP000078492 UP000279307 UP000008820 UP000053097 UP000053825 UP000095300 UP000078200 UP000092444 UP000092445 UP000192223 UP000019118 UP000030742 UP000002282 UP000075883 UP000092460 UP000092443 UP000007801 UP000037069 UP000091820 UP000000803 UP000092553 UP000008792 UP000001070
UP000000311 UP000053105 UP000008237 UP000075809 UP000078542 UP000005205 UP000009192 UP000005203 UP000095301 UP000078541 UP000078492 UP000279307 UP000008820 UP000053097 UP000053825 UP000095300 UP000078200 UP000092444 UP000092445 UP000192223 UP000019118 UP000030742 UP000002282 UP000075883 UP000092460 UP000092443 UP000007801 UP000037069 UP000091820 UP000000803 UP000092553 UP000008792 UP000001070
Interpro
IPR013912
Adenylate_cyclase-assoc_CAP_C
+ More
IPR017901 C-CAP_CF_C-like
IPR018106 CAP_CS_N
IPR003124 WH2_dom
IPR028417 CAP_CS_C
IPR013992 Adenylate_cyclase-assoc_CAP_N
IPR036223 CAP_C_sf
IPR001837 Adenylate_cyclase-assoc_CAP
IPR006599 CARP_motif
IPR036222 CAP_N_sf
IPR016098 CAP/MinC_C
IPR004600 TFIIH_Tfb4/GTF2H3
IPR036465 vWFA_dom_sf
IPR017901 C-CAP_CF_C-like
IPR018106 CAP_CS_N
IPR003124 WH2_dom
IPR028417 CAP_CS_C
IPR013992 Adenylate_cyclase-assoc_CAP_N
IPR036223 CAP_C_sf
IPR001837 Adenylate_cyclase-assoc_CAP
IPR006599 CARP_motif
IPR036222 CAP_N_sf
IPR016098 CAP/MinC_C
IPR004600 TFIIH_Tfb4/GTF2H3
IPR036465 vWFA_dom_sf
Gene 3D
ProteinModelPortal
A0A2A4IVW7
A0A194QUR9
A0A2W1BJX1
A0A212F1V6
A0A1E1WD75
A0A194PPP8
+ More
D6W6L7 A0A154P1C4 A0A1L8E5T3 A0A1L8E5U4 A0A1L8E573 E2ARB8 A0A1B6DH10 A0A0M9A6C6 A0A1B6C4R3 E2BWC7 A0A151XC21 A0A1B6IJ81 A0A1B6FWV4 A0A151IQD3 A0A336KDK4 A0A158NGD4 B4KEU8 A0A087ZPC0 A0A1I8NBN4 A0A195F1A6 A0A195E7R5 T1PCY9 A0A336KDG3 W8BFG0 T1PLK0 A0A1B6F8A1 A0A3L8D7K4 Q16K93 A0A1B6JNG9 A0A026VVZ0 A0A1Q3FG29 A0A0L7R3P1 A0A1Q3FG63 W8BT17 A0A0Q9XIS5 A0A1I8PFE4 U5EUZ2 A0A1I8PF50 A0A023EVL8 A0A034VE93 A0A0T6B7Q4 A0A0P6K160 A0A0A9W4T0 A0A034VGY7 A0A1A9UTX7 A0A034VGB4 A0A1B0FML2 A0A1A9Z5Y4 A0A1Y1LPT5 A0A1W4X8E2 V5I8H4 A0A224XIV7 A0A2M4A5S5 A0A0P4VL56 J3JUS4 A0A2M4A5M6 A0A2M4A5M3 A0A2M4A5W1 A0A2M4A5I8 A0A0C9Q702 A0A0V0G8D7 A0A1Q3FFZ7 A0A1Q3FGF1 A0A1Q3FGH8 A0A0K8VLI6 A0A1L8EBC2 A0A0K8UJZ0 A0A1I8PFE1 A0A0R1DII5 A0A1L8EBF1 A0A1L8EBC1 A0A034VGZ2 A0A1Q3FG80 A0A1L8EBS0 A0A1Q3FFY7 A0A182MQE0 A0A1B0BDT3 A0A1A9XYS8 A0A0P8ZET0 A0A023F3J3 A0A0L0CFZ8 A0A1Q3FG11 A0A0R1DIJ5 A0A1Q3FFU7 A0A1A9WUC6 Q7KN55 Q9VPX7 A0A069DT58 A0A0M4E8Z6 A0A0Q9WCU9 A0A0Q9WNI2 A0A1S4H405 B4JDF4
D6W6L7 A0A154P1C4 A0A1L8E5T3 A0A1L8E5U4 A0A1L8E573 E2ARB8 A0A1B6DH10 A0A0M9A6C6 A0A1B6C4R3 E2BWC7 A0A151XC21 A0A1B6IJ81 A0A1B6FWV4 A0A151IQD3 A0A336KDK4 A0A158NGD4 B4KEU8 A0A087ZPC0 A0A1I8NBN4 A0A195F1A6 A0A195E7R5 T1PCY9 A0A336KDG3 W8BFG0 T1PLK0 A0A1B6F8A1 A0A3L8D7K4 Q16K93 A0A1B6JNG9 A0A026VVZ0 A0A1Q3FG29 A0A0L7R3P1 A0A1Q3FG63 W8BT17 A0A0Q9XIS5 A0A1I8PFE4 U5EUZ2 A0A1I8PF50 A0A023EVL8 A0A034VE93 A0A0T6B7Q4 A0A0P6K160 A0A0A9W4T0 A0A034VGY7 A0A1A9UTX7 A0A034VGB4 A0A1B0FML2 A0A1A9Z5Y4 A0A1Y1LPT5 A0A1W4X8E2 V5I8H4 A0A224XIV7 A0A2M4A5S5 A0A0P4VL56 J3JUS4 A0A2M4A5M6 A0A2M4A5M3 A0A2M4A5W1 A0A2M4A5I8 A0A0C9Q702 A0A0V0G8D7 A0A1Q3FFZ7 A0A1Q3FGF1 A0A1Q3FGH8 A0A0K8VLI6 A0A1L8EBC2 A0A0K8UJZ0 A0A1I8PFE1 A0A0R1DII5 A0A1L8EBF1 A0A1L8EBC1 A0A034VGZ2 A0A1Q3FG80 A0A1L8EBS0 A0A1Q3FFY7 A0A182MQE0 A0A1B0BDT3 A0A1A9XYS8 A0A0P8ZET0 A0A023F3J3 A0A0L0CFZ8 A0A1Q3FG11 A0A0R1DIJ5 A0A1Q3FFU7 A0A1A9WUC6 Q7KN55 Q9VPX7 A0A069DT58 A0A0M4E8Z6 A0A0Q9WCU9 A0A0Q9WNI2 A0A1S4H405 B4JDF4
PDB
1K8F
E-value=1.55212e-41,
Score=427
Ontologies
GO
GO:0003779
GO:0000902
GO:0007010
GO:0007163
GO:0030864
GO:0008179
GO:0008154
GO:0005737
GO:0008360
GO:0042052
GO:0046716
GO:0072499
GO:0008103
GO:0048190
GO:0008407
GO:0006289
GO:0000439
GO:0006355
GO:0048749
GO:0007015
GO:0048477
GO:0005829
GO:0019358
GO:0007018
GO:0030286
GO:0005216
GO:0045211
GO:0006281
GO:0048384
PANTHER
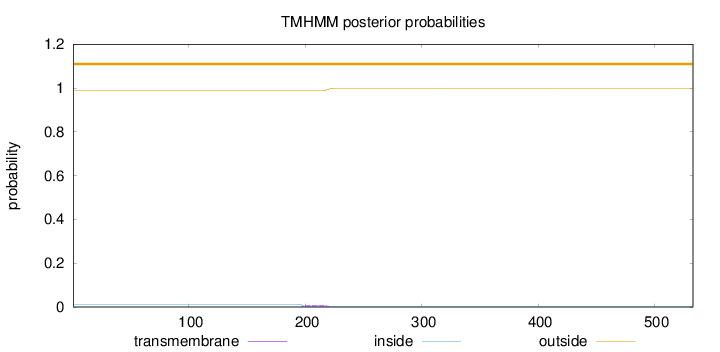
Topology
Length:
533
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19571
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01092
outside
1 - 533
Population Genetic Test Statistics
Pi
225.318678
Theta
175.815085
Tajima's D
0.752453
CLR
0.28416
CSRT
0.587020648967552
Interpretation
Uncertain
