Pre Gene Modal
BGIBMGA003513
Annotation
PREDICTED:_nicotinate_phosphoribosyltransferase_isoform_X2_[Bombyx_mori]
Full name
Nicotinate phosphoribosyltransferase
Location in the cell
Cytoplasmic Reliability : 1.144 PlasmaMembrane Reliability : 1.549
Sequence
CDS
ATGGCTCGACAAAACGGTATTGTGCAACCACTTTTAACTGACCTTTATCAAATAACTATGGCGTATGCATATTGGAAATCAGGAAAAGTAAATGATGTCGCTGTATTTGATTTATTTTTTCGAACAAATCCATTCCAAGGTGAATTCACTATATTTGCTGGATTAGAGGAGTGTATGAAATTTTTAGAAAACTTTCATTATTCTAACAGCGATATCAAATATTTGAAGCAAACATTACCTGAAAACATTGAACCTGAATTTTATTTATATCTCAAAGAACTAACTTGCAAGGACATTATTGTAAGTGCTATTGAAGAAGGATCTGTGGTGTTTCCGAGGGTGCCATTGTTGAGAGTTGAAGGTCCGCTCATTGTGGCACAGCTATTGGAGACCACTCTGCTAACATTAGTCAACTTTGCCAGTTTGATGGCCACGAACGCTGCGAGATATCGGATGGTGGCCGGTAAAAATGTATCCCTACTCGAATTCGGACTACGGAGAGCGCAAGGACCCGATGGAGGGCTATCGGCGTCCAAATATGCGTACATCGGTGGGTTTGACGGGACCAGCAACGTTCTGGCGGGAAAGATGTTCAATATACCGGTAAAGGGCACCCACGCGCATTCGTTCGTTACATCCTTCAGTACTTTAGACGACATACATAGAGTGACGCTGACGCACGCGCAGACACGAGAGCCGTGTAATCTTCTAGAGTCGGCAATGGAATGGCGGAGTCGCGTCTCTACCGTTATCGATGTATCGACAGAGGAATCCAGCGATGGCGAACTGGCCGCCCTAATATCATACGCCATAGCCTTTCCGACGGGTTTCTTGGCTTTAGTGGATACGTACGATGTCAAGAGGAGTGGACTACTGAACTTTTGTGCTGTGGCCCTAGCGTTAAACGATTGTGGCTACAGAGCCGTGGGCATTAGGATCGATAGTGGTGATTTGGCGTATCTATCAGTACTGGCCAGGGAGACGTTCGAAAAGGTCGGGGAAACATTCAACGTCCCTTGGTTCGCTAAACTAACAATCGTCGCATCCAATGATATTAATGAAGAAACCATTCTAAGTTTAAACGAGCAGGGTCATAAAATCGATTGTTTCGGCATCGGAACGCATTTAGTGACATGTCAGCGGCAACCTGCTCTCGGTTGCGTCTACAAGTTGGTCGAAATAAACGGCCAGCCGCGTATAAAGCTCAGCCAGGACGTTGGCAAAGTAACCATACCCGGACACAAAGAGGCATATCGGCTGTTCGGTGCGGATGGACACGCGCTTATAGATTTACTGCAGAGATCTTCGGAACCAGCGCCGGTCGTCGGTCAGAAAGTGCTCTGCCGACATCCGTTCCAAGAGTCGAAACGAGCCTACGTTATCCCCAGCTATGTCGAACATCTTTATAAGATATGCTGGAAAGATGGACGCGTTTGTACGGCACCAAAACCCTTGTCAAAGGTTAGAGAGAGGGTACAGTCGTCTCTAAGGACACTTCGTCAAGACATTAAACGTAACTTGAATCCTACGCCTTATAAGGTGGCCGTCAGCGACGATCTATACAACTTTATCCACGACTTGTGGCTTCAGAATGCACCCATTGGTGAACTCTCATGA
Protein
MARQNGIVQPLLTDLYQITMAYAYWKSGKVNDVAVFDLFFRTNPFQGEFTIFAGLEECMKFLENFHYSNSDIKYLKQTLPENIEPEFYLYLKELTCKDIIVSAIEEGSVVFPRVPLLRVEGPLIVAQLLETTLLTLVNFASLMATNAARYRMVAGKNVSLLEFGLRRAQGPDGGLSASKYAYIGGFDGTSNVLAGKMFNIPVKGTHAHSFVTSFSTLDDIHRVTLTHAQTREPCNLLESAMEWRSRVSTVIDVSTEESSDGELAALISYAIAFPTGFLALVDTYDVKRSGLLNFCAVALALNDCGYRAVGIRIDSGDLAYLSVLARETFEKVGETFNVPWFAKLTIVASNDINEETILSLNEQGHKIDCFGIGTHLVTCQRQPALGCVYKLVEINGQPRIKLSQDVGKVTIPGHKEAYRLFGADGHALIDLLQRSSEPAPVVGQKVLCRHPFQESKRAYVIPSYVEHLYKICWKDGRVCTAPKPLSKVRERVQSSLRTLRQDIKRNLNPTPYKVAVSDDLYNFIHDLWLQNAPIGELS
Summary
Description
Catalyzes the first step in the biosynthesis of NAD from nicotinic acid, the ATP-dependent synthesis of beta-nicotinate D-ribonucleotide from nicotinate and 5-phospho-D-ribose 1-phosphate.
Catalyzes the first step in the biosynthesis of NAD from nicotinic acid, the ATP-dependent synthesis of beta-nicotinate D-ribonucleotide from nicotinate and 5-phospho-D-ribose 1-phosphate. Helps prevent cellular oxidative stress via its role in NAD biosynthesis.
Catalyzes the first step in the biosynthesis of NAD from nicotinic acid, the ATP-dependent synthesis of beta-nicotinate D-ribonucleotide from nicotinate and 5-phospho-D-ribose 1-phosphate. Helps prevent cellular oxidative stress via its role in NAD biosynthesis.
Catalytic Activity
5-phospho-alpha-D-ribose 1-diphosphate + ATP + H2O + nicotinate = ADP + diphosphate + nicotinate beta-D-ribonucleotide + phosphate
Similarity
Belongs to the NAPRTase family.
Keywords
Alternative splicing
Complete proteome
Ligase
Magnesium
Manganese
Metal-binding
Phosphoprotein
Pyridine nucleotide biosynthesis
Reference proteome
Transferase
Feature
chain Nicotinate phosphoribosyltransferase
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4IZS1
A0A1I8Q967
A0A0R3NZC3
A0A2J7QA82
T1PLD8
A0A1I8MF76
+ More
A0A2J7QA86 A0A1B6F2C5 U5EXT3 A0A1B6MND8 A0A1W4V311 A0A0P8XYA0 A0A0J9QVI6 A0A0Q5WI11 A0A0R1DLC4 A0A0A1XAE0 Q9VQX4 B4MTU7 A0A2C9GPV1 A0A1S4GYI1 A0A2J7QA95 A0A1B6FY86 W8B1V5 A0A1B6LIN8 D3TNW0 A0A0Q9W9I5 A0A0Q9XCH2 A0A1L8DZM2 A0A224XAP9 M9PC31 A0A0Q5WLB3 A0A0R1DK39 A0A0V0GCK7 A0A0Q9X0M0 A0A023F4Q3 A0A034VPW0 A0A1Q3FVH5 A0A146L4K1 A0A1B6D5P0 A0A2M3Z6D4 A0A2M3Z6F1 A0A2M4BKB3 Q7PJC3 A0A2M4BKG5 A0A146LCM2 A0A2M4AHH3 E0VUU0 A0A2M4CT37 A0A146LHJ2 A0A1B6D7H7 A0A2J7QA81 A0A0A9WU06 A0A1B6DS14 A0A0A9WIS4 A0A1Y1LHZ7 A0A0A9WLM9 T1J6S6 A0A2H8TLY5 A0A0C9QU29 A0A0A9X2X5 A0A0P4W8R4 A0A162D562 A0A2S2R5I4 A0A1D2NIX7 J9JV22 E9GW48 D6W6L6 A0A087UMM4 A0A1W4W644 N6U0S4 A0A0P6I3J8 A0A0C9RMQ6 A0A2P6K366 U4UUN2 D6X3H3 A0A0J7KP25 A0A1S3GZZ9 A0A2S2PF05 A0A224YUP1 A0A131YLH8 L7MG99 L7MH61 A0A1Z5LBJ1 A0A293LMH8 K1PRY2 A0A131YJ92 A0A2R5L824 A0A0P6DVZ3 A0A0P6CUI7 A0A131YB72 A0A1E1X989 A0A023GL53 A0A2C9JKT3 A0A131XZZ8 B7Q4P8 A0A0P5HNE1 A0A0P5PFC2 A0A0K2TE07 A0A2T7PF69 N6TVQ7
A0A2J7QA86 A0A1B6F2C5 U5EXT3 A0A1B6MND8 A0A1W4V311 A0A0P8XYA0 A0A0J9QVI6 A0A0Q5WI11 A0A0R1DLC4 A0A0A1XAE0 Q9VQX4 B4MTU7 A0A2C9GPV1 A0A1S4GYI1 A0A2J7QA95 A0A1B6FY86 W8B1V5 A0A1B6LIN8 D3TNW0 A0A0Q9W9I5 A0A0Q9XCH2 A0A1L8DZM2 A0A224XAP9 M9PC31 A0A0Q5WLB3 A0A0R1DK39 A0A0V0GCK7 A0A0Q9X0M0 A0A023F4Q3 A0A034VPW0 A0A1Q3FVH5 A0A146L4K1 A0A1B6D5P0 A0A2M3Z6D4 A0A2M3Z6F1 A0A2M4BKB3 Q7PJC3 A0A2M4BKG5 A0A146LCM2 A0A2M4AHH3 E0VUU0 A0A2M4CT37 A0A146LHJ2 A0A1B6D7H7 A0A2J7QA81 A0A0A9WU06 A0A1B6DS14 A0A0A9WIS4 A0A1Y1LHZ7 A0A0A9WLM9 T1J6S6 A0A2H8TLY5 A0A0C9QU29 A0A0A9X2X5 A0A0P4W8R4 A0A162D562 A0A2S2R5I4 A0A1D2NIX7 J9JV22 E9GW48 D6W6L6 A0A087UMM4 A0A1W4W644 N6U0S4 A0A0P6I3J8 A0A0C9RMQ6 A0A2P6K366 U4UUN2 D6X3H3 A0A0J7KP25 A0A1S3GZZ9 A0A2S2PF05 A0A224YUP1 A0A131YLH8 L7MG99 L7MH61 A0A1Z5LBJ1 A0A293LMH8 K1PRY2 A0A131YJ92 A0A2R5L824 A0A0P6DVZ3 A0A0P6CUI7 A0A131YB72 A0A1E1X989 A0A023GL53 A0A2C9JKT3 A0A131XZZ8 B7Q4P8 A0A0P5HNE1 A0A0P5PFC2 A0A0K2TE07 A0A2T7PF69 N6TVQ7
EC Number
6.3.4.21
Pubmed
15632085
17994087
25315136
22936249
17550304
25830018
+ More
10731132 12537572 12537569 12364791 24495485 20353571 12537568 12537573 12537574 16110336 17569856 17569867 25474469 25348373 26823975 14747013 17210077 20566863 25401762 28004739 27289101 21292972 18362917 19820115 23537049 28797301 26830274 25576852 28528879 22992520 28503490 15562597
10731132 12537572 12537569 12364791 24495485 20353571 12537568 12537573 12537574 16110336 17569856 17569867 25474469 25348373 26823975 14747013 17210077 20566863 25401762 28004739 27289101 21292972 18362917 19820115 23537049 28797301 26830274 25576852 28528879 22992520 28503490 15562597
EMBL
NWSH01004297
PCG65251.1
CH379060
KRT03926.1
NEVH01016332
PNF25493.1
+ More
KA649566 AFP64195.1 PNF25494.1 GECZ01025472 GECZ01018863 JAS44297.1 JAS50906.1 GANO01000774 JAB59097.1 GEBQ01002499 JAT37478.1 CH902624 KPU74487.1 CM002910 KMY88026.1 CH954177 KQS70075.1 CM000157 KRJ97245.1 GBXI01014780 GBXI01014645 GBXI01006210 JAC99511.1 JAC99646.1 JAD08082.1 AE014134 AY119468 CH963852 EDW75536.2 APCN01000743 AAAB01008964 PNF25496.1 GECZ01014625 JAS55144.1 GAMC01015552 JAB91003.1 GEBQ01016431 JAT23546.1 EZ423112 ADD19388.1 CH940649 KRF81401.1 CH933807 KRG02580.1 GFDF01002171 JAV11913.1 GFTR01007011 JAW09415.1 AGB92551.1 KQS70074.1 KRJ97246.1 GECL01001005 JAP05119.1 KRF98267.1 GBBI01002320 JAC16392.1 GAKP01013625 GAKP01013621 JAC45331.1 GFDL01003569 JAV31476.1 GDHC01016507 JAQ02122.1 GEDC01016306 JAS20992.1 GGFM01003320 MBW24071.1 GGFM01003319 MBW24070.1 GGFJ01004354 MBW53495.1 AY214334 AAP69612.1 EAA43816.3 GGFJ01004353 MBW53494.1 GDHC01013230 JAQ05399.1 GGFK01006910 MBW40231.1 AAZO01005538 DS235793 EEB17146.1 GGFL01004336 MBW68514.1 GDHC01011068 JAQ07561.1 GEDC01015773 JAS21525.1 PNF25495.1 GBHO01035269 GBRD01016039 JAG08335.1 JAG49787.1 GEDC01008843 JAS28455.1 GBHO01035267 JAG08337.1 GEZM01057006 JAV72511.1 GBHO01035268 JAG08336.1 JH431887 GFXV01003372 MBW15177.1 GBYB01004177 JAG73944.1 GBHO01029588 JAG14016.1 GDRN01064315 JAI64861.1 LRGB01002580 KZS07054.1 GGMS01016052 MBY85255.1 LJIJ01000030 ODN05065.1 ABLF02017331 GL732569 EFX76281.1 KQ971307 EFA10857.2 KK120589 KFM78613.1 APGK01047183 APGK01047184 APGK01047185 KB741077 KB631792 ENN74206.1 ERL86165.1 GDIP01044461 GDIQ01011008 JAM59254.1 JAN83729.1 GBYB01008266 JAG78033.1 MWRG01033973 PRD20752.1 KB632375 ERL93855.1 KQ971374 EEZ97420.1 LBMM01004870 KMQ92026.1 GGMR01015408 MBY28027.1 GFPF01010180 MAA21326.1 GEDV01009195 JAP79362.1 GACK01001808 JAA63226.1 GACK01001882 JAA63152.1 GFJQ02002320 JAW04650.1 GFWV01009968 MAA34697.1 JH815810 EKC24433.1 GEDV01009194 JAP79363.1 GGLE01001502 MBY05628.1 GDIQ01086605 JAN08132.1 GDIQ01088343 JAN06394.1 GEDV01012018 JAP76539.1 GFAC01003602 JAT95586.1 GBBM01000781 JAC34637.1 GEFM01003968 JAP71828.1 ABJB010140890 ABJB010866274 DS857340 EEC13820.1 GDIQ01224849 JAK26876.1 GDIQ01150764 JAL00962.1 HACA01006897 CDW24258.1 PZQS01000004 PVD32044.1 APGK01016134 KB739754 ENN82128.1
KA649566 AFP64195.1 PNF25494.1 GECZ01025472 GECZ01018863 JAS44297.1 JAS50906.1 GANO01000774 JAB59097.1 GEBQ01002499 JAT37478.1 CH902624 KPU74487.1 CM002910 KMY88026.1 CH954177 KQS70075.1 CM000157 KRJ97245.1 GBXI01014780 GBXI01014645 GBXI01006210 JAC99511.1 JAC99646.1 JAD08082.1 AE014134 AY119468 CH963852 EDW75536.2 APCN01000743 AAAB01008964 PNF25496.1 GECZ01014625 JAS55144.1 GAMC01015552 JAB91003.1 GEBQ01016431 JAT23546.1 EZ423112 ADD19388.1 CH940649 KRF81401.1 CH933807 KRG02580.1 GFDF01002171 JAV11913.1 GFTR01007011 JAW09415.1 AGB92551.1 KQS70074.1 KRJ97246.1 GECL01001005 JAP05119.1 KRF98267.1 GBBI01002320 JAC16392.1 GAKP01013625 GAKP01013621 JAC45331.1 GFDL01003569 JAV31476.1 GDHC01016507 JAQ02122.1 GEDC01016306 JAS20992.1 GGFM01003320 MBW24071.1 GGFM01003319 MBW24070.1 GGFJ01004354 MBW53495.1 AY214334 AAP69612.1 EAA43816.3 GGFJ01004353 MBW53494.1 GDHC01013230 JAQ05399.1 GGFK01006910 MBW40231.1 AAZO01005538 DS235793 EEB17146.1 GGFL01004336 MBW68514.1 GDHC01011068 JAQ07561.1 GEDC01015773 JAS21525.1 PNF25495.1 GBHO01035269 GBRD01016039 JAG08335.1 JAG49787.1 GEDC01008843 JAS28455.1 GBHO01035267 JAG08337.1 GEZM01057006 JAV72511.1 GBHO01035268 JAG08336.1 JH431887 GFXV01003372 MBW15177.1 GBYB01004177 JAG73944.1 GBHO01029588 JAG14016.1 GDRN01064315 JAI64861.1 LRGB01002580 KZS07054.1 GGMS01016052 MBY85255.1 LJIJ01000030 ODN05065.1 ABLF02017331 GL732569 EFX76281.1 KQ971307 EFA10857.2 KK120589 KFM78613.1 APGK01047183 APGK01047184 APGK01047185 KB741077 KB631792 ENN74206.1 ERL86165.1 GDIP01044461 GDIQ01011008 JAM59254.1 JAN83729.1 GBYB01008266 JAG78033.1 MWRG01033973 PRD20752.1 KB632375 ERL93855.1 KQ971374 EEZ97420.1 LBMM01004870 KMQ92026.1 GGMR01015408 MBY28027.1 GFPF01010180 MAA21326.1 GEDV01009195 JAP79362.1 GACK01001808 JAA63226.1 GACK01001882 JAA63152.1 GFJQ02002320 JAW04650.1 GFWV01009968 MAA34697.1 JH815810 EKC24433.1 GEDV01009194 JAP79363.1 GGLE01001502 MBY05628.1 GDIQ01086605 JAN08132.1 GDIQ01088343 JAN06394.1 GEDV01012018 JAP76539.1 GFAC01003602 JAT95586.1 GBBM01000781 JAC34637.1 GEFM01003968 JAP71828.1 ABJB010140890 ABJB010866274 DS857340 EEC13820.1 GDIQ01224849 JAK26876.1 GDIQ01150764 JAL00962.1 HACA01006897 CDW24258.1 PZQS01000004 PVD32044.1 APGK01016134 KB739754 ENN82128.1
Proteomes
UP000218220
UP000095300
UP000001819
UP000235965
UP000095301
UP000192221
+ More
UP000007801 UP000008711 UP000002282 UP000000803 UP000007798 UP000075840 UP000008792 UP000009192 UP000007062 UP000009046 UP000076858 UP000094527 UP000007819 UP000000305 UP000007266 UP000054359 UP000192223 UP000019118 UP000030742 UP000036403 UP000085678 UP000005408 UP000076420 UP000001555 UP000245119
UP000007801 UP000008711 UP000002282 UP000000803 UP000007798 UP000075840 UP000008792 UP000009192 UP000007062 UP000009046 UP000076858 UP000094527 UP000007819 UP000000305 UP000007266 UP000054359 UP000192223 UP000019118 UP000030742 UP000036403 UP000085678 UP000005408 UP000076420 UP000001555 UP000245119
Interpro
SUPFAM
SSF51690
SSF51690
Gene 3D
ProteinModelPortal
A0A2A4IZS1
A0A1I8Q967
A0A0R3NZC3
A0A2J7QA82
T1PLD8
A0A1I8MF76
+ More
A0A2J7QA86 A0A1B6F2C5 U5EXT3 A0A1B6MND8 A0A1W4V311 A0A0P8XYA0 A0A0J9QVI6 A0A0Q5WI11 A0A0R1DLC4 A0A0A1XAE0 Q9VQX4 B4MTU7 A0A2C9GPV1 A0A1S4GYI1 A0A2J7QA95 A0A1B6FY86 W8B1V5 A0A1B6LIN8 D3TNW0 A0A0Q9W9I5 A0A0Q9XCH2 A0A1L8DZM2 A0A224XAP9 M9PC31 A0A0Q5WLB3 A0A0R1DK39 A0A0V0GCK7 A0A0Q9X0M0 A0A023F4Q3 A0A034VPW0 A0A1Q3FVH5 A0A146L4K1 A0A1B6D5P0 A0A2M3Z6D4 A0A2M3Z6F1 A0A2M4BKB3 Q7PJC3 A0A2M4BKG5 A0A146LCM2 A0A2M4AHH3 E0VUU0 A0A2M4CT37 A0A146LHJ2 A0A1B6D7H7 A0A2J7QA81 A0A0A9WU06 A0A1B6DS14 A0A0A9WIS4 A0A1Y1LHZ7 A0A0A9WLM9 T1J6S6 A0A2H8TLY5 A0A0C9QU29 A0A0A9X2X5 A0A0P4W8R4 A0A162D562 A0A2S2R5I4 A0A1D2NIX7 J9JV22 E9GW48 D6W6L6 A0A087UMM4 A0A1W4W644 N6U0S4 A0A0P6I3J8 A0A0C9RMQ6 A0A2P6K366 U4UUN2 D6X3H3 A0A0J7KP25 A0A1S3GZZ9 A0A2S2PF05 A0A224YUP1 A0A131YLH8 L7MG99 L7MH61 A0A1Z5LBJ1 A0A293LMH8 K1PRY2 A0A131YJ92 A0A2R5L824 A0A0P6DVZ3 A0A0P6CUI7 A0A131YB72 A0A1E1X989 A0A023GL53 A0A2C9JKT3 A0A131XZZ8 B7Q4P8 A0A0P5HNE1 A0A0P5PFC2 A0A0K2TE07 A0A2T7PF69 N6TVQ7
A0A2J7QA86 A0A1B6F2C5 U5EXT3 A0A1B6MND8 A0A1W4V311 A0A0P8XYA0 A0A0J9QVI6 A0A0Q5WI11 A0A0R1DLC4 A0A0A1XAE0 Q9VQX4 B4MTU7 A0A2C9GPV1 A0A1S4GYI1 A0A2J7QA95 A0A1B6FY86 W8B1V5 A0A1B6LIN8 D3TNW0 A0A0Q9W9I5 A0A0Q9XCH2 A0A1L8DZM2 A0A224XAP9 M9PC31 A0A0Q5WLB3 A0A0R1DK39 A0A0V0GCK7 A0A0Q9X0M0 A0A023F4Q3 A0A034VPW0 A0A1Q3FVH5 A0A146L4K1 A0A1B6D5P0 A0A2M3Z6D4 A0A2M3Z6F1 A0A2M4BKB3 Q7PJC3 A0A2M4BKG5 A0A146LCM2 A0A2M4AHH3 E0VUU0 A0A2M4CT37 A0A146LHJ2 A0A1B6D7H7 A0A2J7QA81 A0A0A9WU06 A0A1B6DS14 A0A0A9WIS4 A0A1Y1LHZ7 A0A0A9WLM9 T1J6S6 A0A2H8TLY5 A0A0C9QU29 A0A0A9X2X5 A0A0P4W8R4 A0A162D562 A0A2S2R5I4 A0A1D2NIX7 J9JV22 E9GW48 D6W6L6 A0A087UMM4 A0A1W4W644 N6U0S4 A0A0P6I3J8 A0A0C9RMQ6 A0A2P6K366 U4UUN2 D6X3H3 A0A0J7KP25 A0A1S3GZZ9 A0A2S2PF05 A0A224YUP1 A0A131YLH8 L7MG99 L7MH61 A0A1Z5LBJ1 A0A293LMH8 K1PRY2 A0A131YJ92 A0A2R5L824 A0A0P6DVZ3 A0A0P6CUI7 A0A131YB72 A0A1E1X989 A0A023GL53 A0A2C9JKT3 A0A131XZZ8 B7Q4P8 A0A0P5HNE1 A0A0P5PFC2 A0A0K2TE07 A0A2T7PF69 N6TVQ7
PDB
4YUB
E-value=6.85462e-139,
Score=1267
Ontologies
PATHWAY
GO
PANTHER
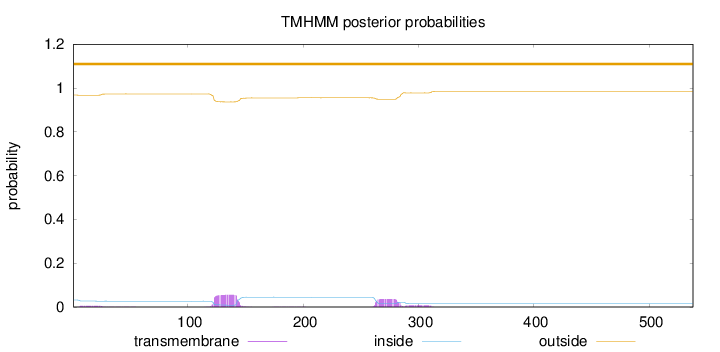
Topology
Length:
538
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.2295
Exp number, first 60 AAs:
0.11637
Total prob of N-in:
0.03233
outside
1 - 538
Population Genetic Test Statistics
Pi
219.077855
Theta
181.222258
Tajima's D
0.59827
CLR
1.07407
CSRT
0.544172791360432
Interpretation
Uncertain
