Gene
KWMTBOMO02822
Annotation
THAP_domain-containing_protein_9_[Crassostrea_gigas]
Location in the cell
Nuclear Reliability : 2.916
Sequence
CDS
ATGAAACCAGTTTCTGAAGAAACAAAAAGTAGAGAACAAAGGTTACAAACTCGTAACCTTAAAAAACAGCTTTTTAGCCTGCCCAACACTTCCCAATACTATGAAGATATAGTACCCCTTGAAGAACACAGCAATGTGCATTTTGATTCAAATGTCTCTCAAGAAATTCTAATAGAAAATAATAACTCTAGTATAGATGAAACAATTACAAGGCATCATATTACTATTGGAACTCAAACATCCAATATATTAAAATTGTTCTCTACAGAGCTATTACTTGCAGACGATGAGACAGTGCAATACTACACCGGGTTGGAAACAGCATCTAAGTTTTCATTAGTTTTAATCACACTTTTGCCAATGTCAAATGATATAAGGTATCGCTGGAGTAGAGTGGTCGGCATATCAATAGAAGATCAATTTTTAATGTTACTAATTAAGCTTAGAAGAAACAACAGATTTTGA
Protein
MKPVSEETKSREQRLQTRNLKKQLFSLPNTSQYYEDIVPLEEHSNVHFDSNVSQEILIENNNSSIDETITRHHITIGTQTSNILKLFSTELLLADDETVQYYTGLETASKFSLVLITLLPMSNDIRYRWSRVVGISIEDQFLMLLIKLRRNNRF
Summary
Uniprot
Pubmed
EMBL
Proteomes
Interpro
Gene 3D
ProteinModelPortal
Ontologies
GO
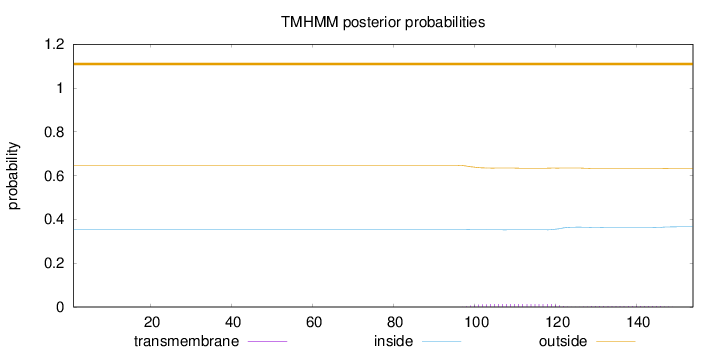
Topology
Length:
154
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.37804
Exp number, first 60 AAs:
0
Total prob of N-in:
0.35333
outside
1 - 154
Population Genetic Test Statistics
Pi
289.799987
Theta
220.002222
Tajima's D
0.988023
CLR
0.156492
CSRT
0.660216989150542
Interpretation
Uncertain
