Gene
KWMTBOMO02819 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003512
Annotation
glucosidase_precursor_[Bombyx_mori]
Location in the cell
Lysosomal Reliability : 1.581
Sequence
CDS
ATGGCTTGGTTAACAACTCTCTCAATACTGGCGGTGTGCCATACAGGATTAGCCGCATACACTAAGTTTCCTGACGGCTTTACATTCGGTGTGGCAACAGCTTCCCATCAAATCGAGGGTGCTTGGAATGTCAGCGGGAAATCCGAAAATGTGTGGGATCGTTTAACCCACACCCGCCCTGAGATGATAGCTGATGGAACCAACGGAGATGTTGCATGTGATTCGTACCATCGTTACCTTGAGGATGTGGAGGAGCTCACTTACCTTGGCGTTGATTTTTACCGGTTCTCGCTTTCCTGGTCACGGATATTGCCAACTGGTTTCTCGGATCACGTGAATCCTGACGGAATTCGGTACTATAACGCTCTCCTCGATGCTCTTGCCGAGAAAAATATCGAGCCTCTTGTCACACTCTTTCACTGGGACTTACCTCAGTCATTACAAGACCTCGGAGGCTGGACTAATTCCAAAATGGTTGATTATTTCCGTGATTACTCCGACGTGTGCTACAGGGAATTCGGTGACAAGATTAAGTCGTGGATTACTATTAACGAACCCTACGAGGTATGCGAAGATGCCTATGGCGATATTAAGAAGGCCCCCGCTTTGGACAGCCATGGCATCGGCAACTATCTATGCAGTGATAATTTACTGAAGGCTCACGCTGAATCTTATCATCTCTACAACGAAAAGTACCGCCCTACTCAAAACGGTACAGTTATGATTTCTATTAATTCGATCTGGTATGAACCTAGCGATCCGGAAAATGCAGAGCAGGTTGCGTTGGCTGAAACTGCCAATCAGTTTAAATTCGGTTGGTTTGCACATCCAATATTTACCAACGAAGGTGGTTATCCTGAAGTTATGATTGAAAATGTCGCAAAGCAAAGCGCAGCTGAAGGTTTCAGCAAGCCCCGATTAGAACAGTTTGATGATTATTGGATTCAAAGGATTAAGGGAACTTCCGATTTTCTGGGTATTAACCACTATGCTACACACTTGATTACTGGCCCCGGAATCGACCCAATAGCTAAATCCCCGTCCTGGTTGAAGGATACTGGCTCCATTACTAGTCTAGAAGTCGGCGGAGATTCAGCTTCGGAGTGGCTTAGGGTGGTGCCGACTGGCTTCGCTAATCTGCTACGTTGGTGCAAGAGCTCGTACAATGACCCACCGATTTACATAACTGAAAATGGATTTTCCGATCGCGGTACCTTGCAAGACTACGGACGCATCCAGTATTACAACGACTATCTGTCTGCGATATTGGATGTTATCTACGATGATGGCGTGAGAGTCCTGGGTTACACCGCGTGGACTCTTATGGACAATTTCGAATGGCGTGCTGGATTCACGGAACCTTTTGGCTTCTACCACGTGGACATCACCGATCCGGACCTACCGAGGACTCCTAAACTCTCAGCAGATTATTATCGCCAGCTCGTCGCCAACCGCGAACTGCCCCAGGACGAGCGTTTTAAGGACCCAGCTGTGAGGAAATAA
Protein
MAWLTTLSILAVCHTGLAAYTKFPDGFTFGVATASHQIEGAWNVSGKSENVWDRLTHTRPEMIADGTNGDVACDSYHRYLEDVEELTYLGVDFYRFSLSWSRILPTGFSDHVNPDGIRYYNALLDALAEKNIEPLVTLFHWDLPQSLQDLGGWTNSKMVDYFRDYSDVCYREFGDKIKSWITINEPYEVCEDAYGDIKKAPALDSHGIGNYLCSDNLLKAHAESYHLYNEKYRPTQNGTVMISINSIWYEPSDPENAEQVALAETANQFKFGWFAHPIFTNEGGYPEVMIENVAKQSAAEGFSKPRLEQFDDYWIQRIKGTSDFLGINHYATHLITGPGIDPIAKSPSWLKDTGSITSLEVGGDSASEWLRVVPTGFANLLRWCKSSYNDPPIYITENGFSDRGTLQDYGRIQYYNDYLSAILDVIYDDGVRVLGYTAWTLMDNFEWRAGFTEPFGFYHVDITDPDLPRTPKLSADYYRQLVANRELPQDERFKDPAVRK
Summary
Similarity
Belongs to the glycosyl hydrolase 1 family.
Uniprot
H9J1X5
Q86D78
A0A194QYR8
A0A194PNC9
A0A2H1V2B7
A0A2A4IZK8
+ More
A0A212F1Y7 A0A2W1BJC8 A0A0L7L473 A0A1W4W814 Q16WF2 A0A182HAK7 A0A1S4G3W5 Q16ET6 A0A0T6B7H1 G9F9G9 B0WNN7 A0A194R360 A0A1W4WJI8 A0A194PMT2 A0A2J7PTJ0 A0A0B5EDK8 A0A1B6D5W8 A0A0K8TVD7 A0A1Y9GKT0 A0A224XNE6 Q5TQY8 V5GZG1 A0A1W4XAC4 A0A1W4WIA6 A0A182U0B4 A0A1Y9IRY8 D6X3T6 T1HZP9 A0A1B0C2P0 R4G3S4 A0A1A9XR01 B4KXZ5 A0A0B5EP46 A0A182YP84 A0A2J7PKK0 T1HZR0 A0A219WGQ6 A0A1Y9J0B2 A0A182QI13 A0A1A9W4X6 B4PKA3 A0A1L8DQN1 A0A1Y9GLV6 A0A2H1VWC6 Q7Q5I1 A0A1A9UP13 B4QMY1 Q9VV98 V5I949 A0A194R3H3 A0A182G2R2 A0A1A9ZLX0 B3NDI0 A0A1Y9IW73 W5JKR6 A0A0B5EM06 A0A1Y9IS68 A0A1W4UV58 A0A0J9RWT3 A0A1W4W9R7 D6WPV3 A0A1Q3FR87 B4H7D1 H9IZH5 A0A194PPG4 A0A1B6FU71 A0A1W1EGM5 A0A0T6B9C6 B4HK62 Q29DT6 B4MM56 A0A1L8DQS5 A0A1L8DR18 G9F9I3 Q8T0W7 A0A0K8TVF8 B3M4E4 A0A2A4JPS5 A0A182LL23 A0A1B6EPT5 A0A182HAK9 A0A0A1XAG3 W5JK33 A0A2M3ZET6 A0A1B6G4E7 A0A1Y9IW90 A0A1B6FXZ7 A0A194PPG9 A0A182G2R0 A0A0S7EEV2 A0A1B6GBB8 A0A2M4BKN2
A0A212F1Y7 A0A2W1BJC8 A0A0L7L473 A0A1W4W814 Q16WF2 A0A182HAK7 A0A1S4G3W5 Q16ET6 A0A0T6B7H1 G9F9G9 B0WNN7 A0A194R360 A0A1W4WJI8 A0A194PMT2 A0A2J7PTJ0 A0A0B5EDK8 A0A1B6D5W8 A0A0K8TVD7 A0A1Y9GKT0 A0A224XNE6 Q5TQY8 V5GZG1 A0A1W4XAC4 A0A1W4WIA6 A0A182U0B4 A0A1Y9IRY8 D6X3T6 T1HZP9 A0A1B0C2P0 R4G3S4 A0A1A9XR01 B4KXZ5 A0A0B5EP46 A0A182YP84 A0A2J7PKK0 T1HZR0 A0A219WGQ6 A0A1Y9J0B2 A0A182QI13 A0A1A9W4X6 B4PKA3 A0A1L8DQN1 A0A1Y9GLV6 A0A2H1VWC6 Q7Q5I1 A0A1A9UP13 B4QMY1 Q9VV98 V5I949 A0A194R3H3 A0A182G2R2 A0A1A9ZLX0 B3NDI0 A0A1Y9IW73 W5JKR6 A0A0B5EM06 A0A1Y9IS68 A0A1W4UV58 A0A0J9RWT3 A0A1W4W9R7 D6WPV3 A0A1Q3FR87 B4H7D1 H9IZH5 A0A194PPG4 A0A1B6FU71 A0A1W1EGM5 A0A0T6B9C6 B4HK62 Q29DT6 B4MM56 A0A1L8DQS5 A0A1L8DR18 G9F9I3 Q8T0W7 A0A0K8TVF8 B3M4E4 A0A2A4JPS5 A0A182LL23 A0A1B6EPT5 A0A182HAK9 A0A0A1XAG3 W5JK33 A0A2M3ZET6 A0A1B6G4E7 A0A1Y9IW90 A0A1B6FXZ7 A0A194PPG9 A0A182G2R0 A0A0S7EEV2 A0A1B6GBB8 A0A2M4BKN2
Pubmed
19121390
15970451
26354079
22118469
28756777
26227816
+ More
17510324 26483478 25596091 12364791 14747013 17210077 18362917 19820115 17994087 25244985 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 22936249 28680679 15632085 12429120 20682343 22751668 20966253 25830018
17510324 26483478 25596091 12364791 14747013 17210077 18362917 19820115 17994087 25244985 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 22936249 28680679 15632085 12429120 20682343 22751668 20966253 25830018
EMBL
BABH01007773
BABH01007774
AY272037
AAP13852.1
KQ461150
KPJ08731.1
+ More
KQ459597 KPI94941.1 ODYU01000355 SOQ34988.1 NWSH01004297 PCG65255.1 AGBW02010786 OWR47723.1 KZ150007 PZC75182.1 JTDY01003021 KOB70293.1 CH477568 EAT38910.1 JXUM01031352 KQ560919 KXJ80348.1 CH478582 EAT32750.1 LJIG01009590 KRT82775.1 JN033714 AEW46867.1 DS232013 EDS31873.1 KQ460779 KPJ12243.1 KQ459598 KPI94627.1 NEVH01021237 PNF19653.1 KP068696 AJE75667.1 GEDC01016220 JAS21078.1 GBUI01000002 JAI18233.1 APCN01002262 GFTR01006753 JAW09673.1 AAAB01008960 EAL40075.2 GALX01002583 JAB65883.1 KQ971326 EEZ97373.2 ACPB03021503 JXJN01024604 JXJN01024605 JXJN01024606 GAHY01001965 JAA75545.1 CH933809 EDW18697.1 KP068700 AJE75671.1 NEVH01024540 PNF16839.1 KT992464 AOY34571.1 AXCN02000455 CM000159 EDW94801.1 GFDF01005306 JAV08778.1 ODYU01004836 SOQ45125.1 EAA11668.3 CM000363 EDX10773.1 AE014296 AY069733 AAF49418.2 AAL39878.1 GALX01003555 JAB64911.1 KPJ12242.1 JXUM01040979 KQ561266 KXJ79157.1 CH954178 EDV52042.2 ADMH02000892 ETN64736.1 KP068694 AJE75665.1 CM002912 KMZ00102.1 KQ971354 EFA06168.1 GFDL01004884 JAV30161.1 CH479217 EDW33737.1 BABH01014157 KPI94624.1 GECZ01016080 JAS53689.1 LT635664 SGZ49394.1 LJIG01003002 KRT83938.1 CH480815 EDW41803.1 CH379070 EAL30328.1 CH963847 EDW73065.2 GFDF01005305 JAV08779.1 GFDF01005304 JAV08780.1 JN033728 AEW46881.1 AB073638 BAB91145.1 GBUI01000004 JAI18231.1 CH902618 EDV40438.1 NWSH01000819 PCG74065.1 GECZ01029831 JAS39938.1 JXUM01031354 KXJ80350.1 GBXI01006185 JAD08107.1 ETN64737.1 GGFM01006254 MBW27005.1 GECZ01012459 JAS57310.1 GECZ01014768 JAS55001.1 KPI94629.1 JXUM01040976 KXJ79155.1 GBUH01000006 JAO03636.1 GECZ01010057 JAS59712.1 GGFJ01004382 MBW53523.1
KQ459597 KPI94941.1 ODYU01000355 SOQ34988.1 NWSH01004297 PCG65255.1 AGBW02010786 OWR47723.1 KZ150007 PZC75182.1 JTDY01003021 KOB70293.1 CH477568 EAT38910.1 JXUM01031352 KQ560919 KXJ80348.1 CH478582 EAT32750.1 LJIG01009590 KRT82775.1 JN033714 AEW46867.1 DS232013 EDS31873.1 KQ460779 KPJ12243.1 KQ459598 KPI94627.1 NEVH01021237 PNF19653.1 KP068696 AJE75667.1 GEDC01016220 JAS21078.1 GBUI01000002 JAI18233.1 APCN01002262 GFTR01006753 JAW09673.1 AAAB01008960 EAL40075.2 GALX01002583 JAB65883.1 KQ971326 EEZ97373.2 ACPB03021503 JXJN01024604 JXJN01024605 JXJN01024606 GAHY01001965 JAA75545.1 CH933809 EDW18697.1 KP068700 AJE75671.1 NEVH01024540 PNF16839.1 KT992464 AOY34571.1 AXCN02000455 CM000159 EDW94801.1 GFDF01005306 JAV08778.1 ODYU01004836 SOQ45125.1 EAA11668.3 CM000363 EDX10773.1 AE014296 AY069733 AAF49418.2 AAL39878.1 GALX01003555 JAB64911.1 KPJ12242.1 JXUM01040979 KQ561266 KXJ79157.1 CH954178 EDV52042.2 ADMH02000892 ETN64736.1 KP068694 AJE75665.1 CM002912 KMZ00102.1 KQ971354 EFA06168.1 GFDL01004884 JAV30161.1 CH479217 EDW33737.1 BABH01014157 KPI94624.1 GECZ01016080 JAS53689.1 LT635664 SGZ49394.1 LJIG01003002 KRT83938.1 CH480815 EDW41803.1 CH379070 EAL30328.1 CH963847 EDW73065.2 GFDF01005305 JAV08779.1 GFDF01005304 JAV08780.1 JN033728 AEW46881.1 AB073638 BAB91145.1 GBUI01000004 JAI18231.1 CH902618 EDV40438.1 NWSH01000819 PCG74065.1 GECZ01029831 JAS39938.1 JXUM01031354 KXJ80350.1 GBXI01006185 JAD08107.1 ETN64737.1 GGFM01006254 MBW27005.1 GECZ01012459 JAS57310.1 GECZ01014768 JAS55001.1 KPI94629.1 JXUM01040976 KXJ79155.1 GBUH01000006 JAO03636.1 GECZ01010057 JAS59712.1 GGFJ01004382 MBW53523.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
+ More
UP000192223 UP000008820 UP000069940 UP000249989 UP000002320 UP000235965 UP000075840 UP000007062 UP000075902 UP000075903 UP000007266 UP000015103 UP000092460 UP000092443 UP000009192 UP000076408 UP000076407 UP000075886 UP000091820 UP000002282 UP000078200 UP000000304 UP000000803 UP000092445 UP000008711 UP000075920 UP000000673 UP000192221 UP000008744 UP000001292 UP000001819 UP000007798 UP000007801 UP000075882
UP000192223 UP000008820 UP000069940 UP000249989 UP000002320 UP000235965 UP000075840 UP000007062 UP000075902 UP000075903 UP000007266 UP000015103 UP000092460 UP000092443 UP000009192 UP000076408 UP000076407 UP000075886 UP000091820 UP000002282 UP000078200 UP000000304 UP000000803 UP000092445 UP000008711 UP000075920 UP000000673 UP000192221 UP000008744 UP000001292 UP000001819 UP000007798 UP000007801 UP000075882
Pfam
PF00232 Glyco_hydro_1
Interpro
SUPFAM
SSF51445
SSF51445
ProteinModelPortal
H9J1X5
Q86D78
A0A194QYR8
A0A194PNC9
A0A2H1V2B7
A0A2A4IZK8
+ More
A0A212F1Y7 A0A2W1BJC8 A0A0L7L473 A0A1W4W814 Q16WF2 A0A182HAK7 A0A1S4G3W5 Q16ET6 A0A0T6B7H1 G9F9G9 B0WNN7 A0A194R360 A0A1W4WJI8 A0A194PMT2 A0A2J7PTJ0 A0A0B5EDK8 A0A1B6D5W8 A0A0K8TVD7 A0A1Y9GKT0 A0A224XNE6 Q5TQY8 V5GZG1 A0A1W4XAC4 A0A1W4WIA6 A0A182U0B4 A0A1Y9IRY8 D6X3T6 T1HZP9 A0A1B0C2P0 R4G3S4 A0A1A9XR01 B4KXZ5 A0A0B5EP46 A0A182YP84 A0A2J7PKK0 T1HZR0 A0A219WGQ6 A0A1Y9J0B2 A0A182QI13 A0A1A9W4X6 B4PKA3 A0A1L8DQN1 A0A1Y9GLV6 A0A2H1VWC6 Q7Q5I1 A0A1A9UP13 B4QMY1 Q9VV98 V5I949 A0A194R3H3 A0A182G2R2 A0A1A9ZLX0 B3NDI0 A0A1Y9IW73 W5JKR6 A0A0B5EM06 A0A1Y9IS68 A0A1W4UV58 A0A0J9RWT3 A0A1W4W9R7 D6WPV3 A0A1Q3FR87 B4H7D1 H9IZH5 A0A194PPG4 A0A1B6FU71 A0A1W1EGM5 A0A0T6B9C6 B4HK62 Q29DT6 B4MM56 A0A1L8DQS5 A0A1L8DR18 G9F9I3 Q8T0W7 A0A0K8TVF8 B3M4E4 A0A2A4JPS5 A0A182LL23 A0A1B6EPT5 A0A182HAK9 A0A0A1XAG3 W5JK33 A0A2M3ZET6 A0A1B6G4E7 A0A1Y9IW90 A0A1B6FXZ7 A0A194PPG9 A0A182G2R0 A0A0S7EEV2 A0A1B6GBB8 A0A2M4BKN2
A0A212F1Y7 A0A2W1BJC8 A0A0L7L473 A0A1W4W814 Q16WF2 A0A182HAK7 A0A1S4G3W5 Q16ET6 A0A0T6B7H1 G9F9G9 B0WNN7 A0A194R360 A0A1W4WJI8 A0A194PMT2 A0A2J7PTJ0 A0A0B5EDK8 A0A1B6D5W8 A0A0K8TVD7 A0A1Y9GKT0 A0A224XNE6 Q5TQY8 V5GZG1 A0A1W4XAC4 A0A1W4WIA6 A0A182U0B4 A0A1Y9IRY8 D6X3T6 T1HZP9 A0A1B0C2P0 R4G3S4 A0A1A9XR01 B4KXZ5 A0A0B5EP46 A0A182YP84 A0A2J7PKK0 T1HZR0 A0A219WGQ6 A0A1Y9J0B2 A0A182QI13 A0A1A9W4X6 B4PKA3 A0A1L8DQN1 A0A1Y9GLV6 A0A2H1VWC6 Q7Q5I1 A0A1A9UP13 B4QMY1 Q9VV98 V5I949 A0A194R3H3 A0A182G2R2 A0A1A9ZLX0 B3NDI0 A0A1Y9IW73 W5JKR6 A0A0B5EM06 A0A1Y9IS68 A0A1W4UV58 A0A0J9RWT3 A0A1W4W9R7 D6WPV3 A0A1Q3FR87 B4H7D1 H9IZH5 A0A194PPG4 A0A1B6FU71 A0A1W1EGM5 A0A0T6B9C6 B4HK62 Q29DT6 B4MM56 A0A1L8DQS5 A0A1L8DR18 G9F9I3 Q8T0W7 A0A0K8TVF8 B3M4E4 A0A2A4JPS5 A0A182LL23 A0A1B6EPT5 A0A182HAK9 A0A0A1XAG3 W5JK33 A0A2M3ZET6 A0A1B6G4E7 A0A1Y9IW90 A0A1B6FXZ7 A0A194PPG9 A0A182G2R0 A0A0S7EEV2 A0A1B6GBB8 A0A2M4BKN2
PDB
3VII
E-value=8.318e-132,
Score=1206
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.983751
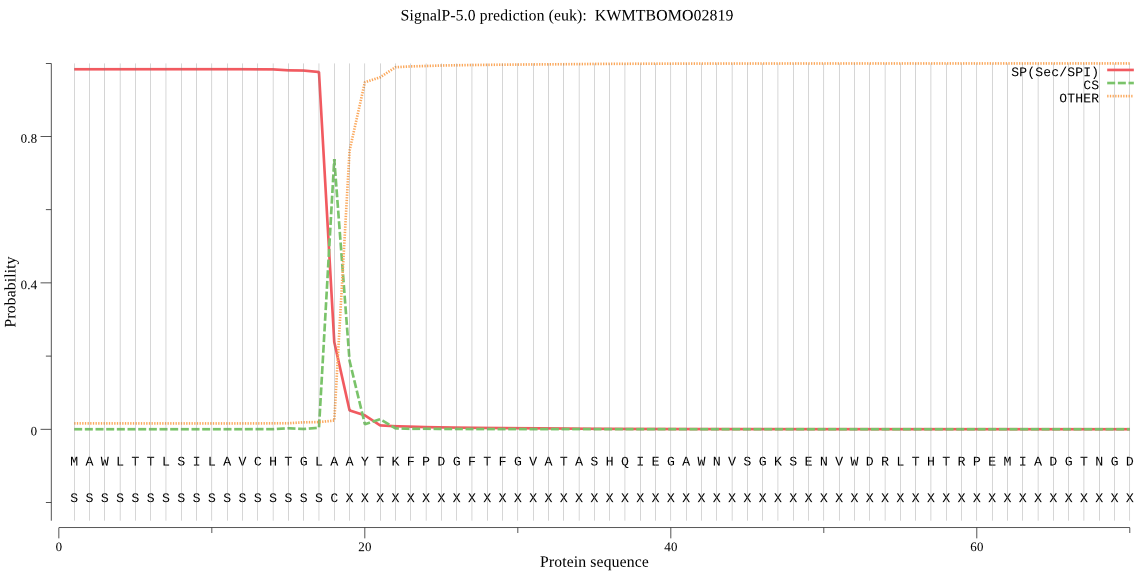
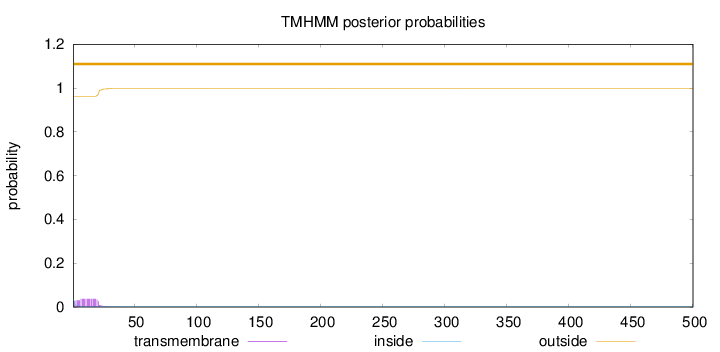
Length:
500
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.73019
Exp number, first 60 AAs:
0.72928
Total prob of N-in:
0.03797
outside
1 - 500
Population Genetic Test Statistics
Pi
211.408805
Theta
211.982832
Tajima's D
0.65986
CLR
0.136264
CSRT
0.55767211639418
Interpretation
Uncertain
