Gene
KWMTBOMO02817
Annotation
cytochrome_P450_CYP4M9_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.822
Sequence
CDS
ATGTGGATGTACTTTATTCTTATTTTACTACTGTTGCTTACAATTCATTTCTTGTTAAATTATAACTACAGAGCTAGATTACTTAGAAGAATTCCGGGACCAAGAGGTTATTTCATCGTTGGAAATGCCTTGGATGTTATTTTGTCACCAGCTGAATTATTCGCATCGACAAGAAAAAATGCAGCCCAATGGCCGAATTTAAATAGATTTTGGTCTTTTGGTATAGGAGCCTTAAATATATACGGACCCGATGAAATTGAGGCTATTATTTCAAGCACACAACATATTACAAAGAGTCCTGTGTACAACTTTTTGAGTGATTGGCTACGAGACGGATTACTTTTGAGTACAGGTACGAAATGGCAAAAACGAAGAAAAATACTGACACCAGCTTTTCATTTCAACATTTTGAAGCAGTTCTGTGTCATTCTGGAAGAAAATAGTCAGCGTTTTACTGAGAATCTAAAGGACACTGAGGGCAAAAGTATTAATGTTGTTCCTGCTATCTCGGAGTACACACTTCATTCAATTTGCGAAACTGCAATGAGAACCCAACTTGGTAGCGAAACGTCTGAAGCAGGAAGATCATATAAGAATGCTATATGTGAGCTCGGAAACCAATTCGTGCACAGACTAGCTAGATTACCTCTCCACAACAACTTCATTTACAACTTGTACACACTGGGAAAGCAAAATAAACACTTAAACATAGTTCATAGCTTCACTAAAAAAGTAATAAAAGATCGCCGTCAATACATAAGAGGGAACGGAGGTAATAATTTTGACGATGAAAAAGACACACAAGCTGACGAACATTCCATTTATTTTAACAAGAAAAAAACAGCTATGTTGGATCTCCTGCTAAAGGCAGAACGAGATGGATTAATTGACGAAATTGGCGTTCAAGAAGAAATCGACACATTCATGTTTGAAGGTCACGACACTACTGCTACCGGATTAACTTATTGCATTATGTTGATAGCAAACCACAAATCAATTCAAGATAAAATAATTGAAGAATTGGATGAAATTTTCGGTGAATCAACGAGAGCCGCTGACATAGAAGATCTATCTAAAATGCGCTATTTAGAAAGGTGCATCAAAGAATCTTTACGATTATATCCACCGGTACCATCAATGGGCCGTATTTTAAGCGAAGAAATCAACCTCAATGGTTACACCGTACCAGCCGGGACCTACTGTCATATTCAAATATTTGACTTACACCGCCGCGAGGATTTGTTCAAAGATCCTCTTGTGTTTGACCCGGACCGTTTCTTACCACATAACACCGAAGGCCGACACCCTTACGCTTACATACCTTTTAGCGCTGGACCCAGAAATTGCATAGGTCAGAAATTTGCCATACTCGAGATGAAGTCTCTTCTGTCTGCTGTATTGCGTCGTTACAATCTGTATCCAATAACCAAACCGGAAGATCTTAAATTTGTTCTAGATCTAGTTTTAAGGACAACGGAACCGGTTCACGTTAGATTTGTGAAGAGAAATAAAGTATAG
Protein
MWMYFILILLLLLTIHFLLNYNYRARLLRRIPGPRGYFIVGNALDVILSPAELFASTRKNAAQWPNLNRFWSFGIGALNIYGPDEIEAIISSTQHITKSPVYNFLSDWLRDGLLLSTGTKWQKRRKILTPAFHFNILKQFCVILEENSQRFTENLKDTEGKSINVVPAISEYTLHSICETAMRTQLGSETSEAGRSYKNAICELGNQFVHRLARLPLHNNFIYNLYTLGKQNKHLNIVHSFTKKVIKDRRQYIRGNGGNNFDDEKDTQADEHSIYFNKKKTAMLDLLLKAERDGLIDEIGVQEEIDTFMFEGHDTTATGLTYCIMLIANHKSIQDKIIEELDEIFGESTRAADIEDLSKMRYLERCIKESLRLYPPVPSMGRILSEEINLNGYTVPAGTYCHIQIFDLHRREDLFKDPLVFDPDRFLPHNTEGRHPYAYIPFSAGPRNCIGQKFAILEMKSLLSAVLRRYNLYPITKPEDLKFVLDLVLRTTEPVHVRFVKRNKV
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
L0N790
A0MNW3
A0MNW4
Q25508
D5L0M1
X5DQR3
+ More
A0A068F0W3 A0A2A4JU89 A0A2W1BPC2 D5L0M0 A0A248QJ75 A5A3F8 L0N4I2 Q2L7F2 L0N5E9 A9QW14 A5HG32 D0FZ08 A0A2H1V2B5 A0A248QFF8 A0A172MA26 X5DT35 C1LZ55 A0A0C5C1N3 A0A248QEJ1 Q8MTC9 Q8MTD0 A0A2W1BJD8 A0A068ETT9 A0A2W1BJV4 V5W638 A0A068EWP1 A0A2A4JUK6 A0A2A4JUC6 A0A194QYS2 A0A248QEI4 A0A286MXM2 A0A286MXM1 A0A194PNP6 A0A212F1V8 A0A1V0D9C5 A0A1E1WBF9 A0A212EXA9 D5L0M2 H9J2E6 A0A1E1WM70 K7IR11 A0A2A4JHP2 A0A1V0D9D1 A0A194PPN8 A0A232EQV5 A0A0C5BXG0 K7J8M6 A0A1W6L1H7 A0A194QUS9 B2BNZ3 A0A2W1BLR4 A0A232EII5 A0A286QUL0 A0A0K1YWX7 J7FIT5 A0A310SVS3 A0A2A4ITL8 A0A1Y1M0Y8 K7J2L1 A0A0K1YW86 A0A158P394 A0A0L7KMS1 A0A232F6N7 Q8ISJ8 A0A068EVK2 A0A0N0PCP3 A0A0K1YWI7 D6X2T3 K7IMP8 A0A068EU59 L0N6M8 A0A0U2DUD7 Q2I0J0 D6X2S9 A0A212F1X5 A0A154PPN0 A0A0L7R2Z5 K7J4W7 L0N716 A0A182F3I3 A0A084WH35 A0A182RIQ7 E2AD32 Q6VAB7 A0A232FKZ8 A0A194QLZ7 A0A195D4T2 A0A1Q3G218 A0A182PE74 A0A2M4BLC8 A0A0K0LBB5 K7J9G3 A0A182I5C4 D6X2T0
A0A068F0W3 A0A2A4JU89 A0A2W1BPC2 D5L0M0 A0A248QJ75 A5A3F8 L0N4I2 Q2L7F2 L0N5E9 A9QW14 A5HG32 D0FZ08 A0A2H1V2B5 A0A248QFF8 A0A172MA26 X5DT35 C1LZ55 A0A0C5C1N3 A0A248QEJ1 Q8MTC9 Q8MTD0 A0A2W1BJD8 A0A068ETT9 A0A2W1BJV4 V5W638 A0A068EWP1 A0A2A4JUK6 A0A2A4JUC6 A0A194QYS2 A0A248QEI4 A0A286MXM2 A0A286MXM1 A0A194PNP6 A0A212F1V8 A0A1V0D9C5 A0A1E1WBF9 A0A212EXA9 D5L0M2 H9J2E6 A0A1E1WM70 K7IR11 A0A2A4JHP2 A0A1V0D9D1 A0A194PPN8 A0A232EQV5 A0A0C5BXG0 K7J8M6 A0A1W6L1H7 A0A194QUS9 B2BNZ3 A0A2W1BLR4 A0A232EII5 A0A286QUL0 A0A0K1YWX7 J7FIT5 A0A310SVS3 A0A2A4ITL8 A0A1Y1M0Y8 K7J2L1 A0A0K1YW86 A0A158P394 A0A0L7KMS1 A0A232F6N7 Q8ISJ8 A0A068EVK2 A0A0N0PCP3 A0A0K1YWI7 D6X2T3 K7IMP8 A0A068EU59 L0N6M8 A0A0U2DUD7 Q2I0J0 D6X2S9 A0A212F1X5 A0A154PPN0 A0A0L7R2Z5 K7J4W7 L0N716 A0A182F3I3 A0A084WH35 A0A182RIQ7 E2AD32 Q6VAB7 A0A232FKZ8 A0A194QLZ7 A0A195D4T2 A0A1Q3G218 A0A182PE74 A0A2M4BLC8 A0A0K0LBB5 K7J9G3 A0A182I5C4 D6X2T0
Pubmed
EMBL
AK289283
BAM73810.1
EF017796
ABK27871.1
EF017797
ABK27872.1
+ More
L38671 AAC21661.1 GU731526 ADE05576.1 KF701163 AHW57333.1 KM016728 AID54880.1 NWSH01000604 PCG75379.1 KZ150007 PZC75157.1 GU731525 ADE05575.1 KX443466 ASO98041.1 EF523434 ABP87671.1 AK289282 BAM73809.1 DQ352137 ABC72321.2 AK289281 BAM73808.1 EU273876 ABX64439.1 EF528484 ABP99018.1 AB498590 BAI45222.1 ODYU01000355 SOQ34985.1 SOQ34986.1 KX443467 ASO98042.1 KX008607 ANC90156.1 KF701164 AHW57334.1 FN356972 CAX94850.1 KP001139 AJN91184.1 KX443468 ASO98043.1 AY113688 AAM54723.1 AY113687 AAM54722.1 PZC75159.1 KM016730 AID54882.1 PZC75158.1 KF668286 AHC02483.1 KM016729 AID54881.1 PCG75378.1 PCG75376.1 KQ461150 KPJ08736.1 KX443469 ASO98044.1 MF684340 ASX93974.1 MF684339 ASX93973.1 KQ459597 KPI94937.1 AGBW02010786 OWR47720.1 KY212038 ARA91602.1 GDQN01006741 JAT84313.1 AGBW02011790 OWR46123.1 GU731527 ADE05577.1 BABH01007780 BABH01007781 GDQN01009628 GDQN01002954 JAT81426.1 JAT88100.1 NWSH01001487 PCG71104.1 KY212037 ARA91601.1 KPI94938.1 NNAY01002731 OXU20676.1 KP001143 AJN91188.1 AAZX01000970 KX609525 ARN17932.1 KPJ08735.1 EF591060 ABU88427.1 KZ150005 PZC75221.1 NNAY01004274 OXU18121.1 KX443461 ASO98036.1 KP859402 AKZ17712.1 JX310089 AFP20600.1 KQ759878 OAD62263.1 NWSH01006615 PCG63327.1 GEZM01042998 JAV79399.1 KP859401 AKZ17711.1 ADTU01007893 ADTU01007894 JTDY01008387 KOB64592.1 NNAY01000868 OXU26140.1 AY063500 AAL48299.1 KM016731 AID54883.1 KQ460479 KPJ14362.1 KP859403 AKZ17713.1 KQ971372 EFA10757.1 KM016732 AID54884.1 PZC75222.1 AK343220 BAM73902.1 KP058451 AKP80586.1 DQ355383 ABC84370.2 EFA10753.1 OWR47721.1 KQ435012 KZC13846.1 KQ414664 KOC65199.1 AK289284 BAM73811.1 ATLV01023783 KE525346 KFB49529.1 GL438658 EFN68668.1 AY345971 AAQ90477.1 NNAY01000058 OXU31426.1 KQ458981 KPJ04476.1 KQ976842 KYN07893.1 GFDL01001167 JAV33878.1 GGFJ01004457 MBW53598.1 KM217034 AIW79997.1 APCN01003852 EFA10754.1
L38671 AAC21661.1 GU731526 ADE05576.1 KF701163 AHW57333.1 KM016728 AID54880.1 NWSH01000604 PCG75379.1 KZ150007 PZC75157.1 GU731525 ADE05575.1 KX443466 ASO98041.1 EF523434 ABP87671.1 AK289282 BAM73809.1 DQ352137 ABC72321.2 AK289281 BAM73808.1 EU273876 ABX64439.1 EF528484 ABP99018.1 AB498590 BAI45222.1 ODYU01000355 SOQ34985.1 SOQ34986.1 KX443467 ASO98042.1 KX008607 ANC90156.1 KF701164 AHW57334.1 FN356972 CAX94850.1 KP001139 AJN91184.1 KX443468 ASO98043.1 AY113688 AAM54723.1 AY113687 AAM54722.1 PZC75159.1 KM016730 AID54882.1 PZC75158.1 KF668286 AHC02483.1 KM016729 AID54881.1 PCG75378.1 PCG75376.1 KQ461150 KPJ08736.1 KX443469 ASO98044.1 MF684340 ASX93974.1 MF684339 ASX93973.1 KQ459597 KPI94937.1 AGBW02010786 OWR47720.1 KY212038 ARA91602.1 GDQN01006741 JAT84313.1 AGBW02011790 OWR46123.1 GU731527 ADE05577.1 BABH01007780 BABH01007781 GDQN01009628 GDQN01002954 JAT81426.1 JAT88100.1 NWSH01001487 PCG71104.1 KY212037 ARA91601.1 KPI94938.1 NNAY01002731 OXU20676.1 KP001143 AJN91188.1 AAZX01000970 KX609525 ARN17932.1 KPJ08735.1 EF591060 ABU88427.1 KZ150005 PZC75221.1 NNAY01004274 OXU18121.1 KX443461 ASO98036.1 KP859402 AKZ17712.1 JX310089 AFP20600.1 KQ759878 OAD62263.1 NWSH01006615 PCG63327.1 GEZM01042998 JAV79399.1 KP859401 AKZ17711.1 ADTU01007893 ADTU01007894 JTDY01008387 KOB64592.1 NNAY01000868 OXU26140.1 AY063500 AAL48299.1 KM016731 AID54883.1 KQ460479 KPJ14362.1 KP859403 AKZ17713.1 KQ971372 EFA10757.1 KM016732 AID54884.1 PZC75222.1 AK343220 BAM73902.1 KP058451 AKP80586.1 DQ355383 ABC84370.2 EFA10753.1 OWR47721.1 KQ435012 KZC13846.1 KQ414664 KOC65199.1 AK289284 BAM73811.1 ATLV01023783 KE525346 KFB49529.1 GL438658 EFN68668.1 AY345971 AAQ90477.1 NNAY01000058 OXU31426.1 KQ458981 KPJ04476.1 KQ976842 KYN07893.1 GFDL01001167 JAV33878.1 GGFJ01004457 MBW53598.1 KM217034 AIW79997.1 APCN01003852 EFA10754.1
Proteomes
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N790
A0MNW3
A0MNW4
Q25508
D5L0M1
X5DQR3
+ More
A0A068F0W3 A0A2A4JU89 A0A2W1BPC2 D5L0M0 A0A248QJ75 A5A3F8 L0N4I2 Q2L7F2 L0N5E9 A9QW14 A5HG32 D0FZ08 A0A2H1V2B5 A0A248QFF8 A0A172MA26 X5DT35 C1LZ55 A0A0C5C1N3 A0A248QEJ1 Q8MTC9 Q8MTD0 A0A2W1BJD8 A0A068ETT9 A0A2W1BJV4 V5W638 A0A068EWP1 A0A2A4JUK6 A0A2A4JUC6 A0A194QYS2 A0A248QEI4 A0A286MXM2 A0A286MXM1 A0A194PNP6 A0A212F1V8 A0A1V0D9C5 A0A1E1WBF9 A0A212EXA9 D5L0M2 H9J2E6 A0A1E1WM70 K7IR11 A0A2A4JHP2 A0A1V0D9D1 A0A194PPN8 A0A232EQV5 A0A0C5BXG0 K7J8M6 A0A1W6L1H7 A0A194QUS9 B2BNZ3 A0A2W1BLR4 A0A232EII5 A0A286QUL0 A0A0K1YWX7 J7FIT5 A0A310SVS3 A0A2A4ITL8 A0A1Y1M0Y8 K7J2L1 A0A0K1YW86 A0A158P394 A0A0L7KMS1 A0A232F6N7 Q8ISJ8 A0A068EVK2 A0A0N0PCP3 A0A0K1YWI7 D6X2T3 K7IMP8 A0A068EU59 L0N6M8 A0A0U2DUD7 Q2I0J0 D6X2S9 A0A212F1X5 A0A154PPN0 A0A0L7R2Z5 K7J4W7 L0N716 A0A182F3I3 A0A084WH35 A0A182RIQ7 E2AD32 Q6VAB7 A0A232FKZ8 A0A194QLZ7 A0A195D4T2 A0A1Q3G218 A0A182PE74 A0A2M4BLC8 A0A0K0LBB5 K7J9G3 A0A182I5C4 D6X2T0
A0A068F0W3 A0A2A4JU89 A0A2W1BPC2 D5L0M0 A0A248QJ75 A5A3F8 L0N4I2 Q2L7F2 L0N5E9 A9QW14 A5HG32 D0FZ08 A0A2H1V2B5 A0A248QFF8 A0A172MA26 X5DT35 C1LZ55 A0A0C5C1N3 A0A248QEJ1 Q8MTC9 Q8MTD0 A0A2W1BJD8 A0A068ETT9 A0A2W1BJV4 V5W638 A0A068EWP1 A0A2A4JUK6 A0A2A4JUC6 A0A194QYS2 A0A248QEI4 A0A286MXM2 A0A286MXM1 A0A194PNP6 A0A212F1V8 A0A1V0D9C5 A0A1E1WBF9 A0A212EXA9 D5L0M2 H9J2E6 A0A1E1WM70 K7IR11 A0A2A4JHP2 A0A1V0D9D1 A0A194PPN8 A0A232EQV5 A0A0C5BXG0 K7J8M6 A0A1W6L1H7 A0A194QUS9 B2BNZ3 A0A2W1BLR4 A0A232EII5 A0A286QUL0 A0A0K1YWX7 J7FIT5 A0A310SVS3 A0A2A4ITL8 A0A1Y1M0Y8 K7J2L1 A0A0K1YW86 A0A158P394 A0A0L7KMS1 A0A232F6N7 Q8ISJ8 A0A068EVK2 A0A0N0PCP3 A0A0K1YWI7 D6X2T3 K7IMP8 A0A068EU59 L0N6M8 A0A0U2DUD7 Q2I0J0 D6X2S9 A0A212F1X5 A0A154PPN0 A0A0L7R2Z5 K7J4W7 L0N716 A0A182F3I3 A0A084WH35 A0A182RIQ7 E2AD32 Q6VAB7 A0A232FKZ8 A0A194QLZ7 A0A195D4T2 A0A1Q3G218 A0A182PE74 A0A2M4BLC8 A0A0K0LBB5 K7J9G3 A0A182I5C4 D6X2T0
PDB
6C94
E-value=4.81102e-74,
Score=708
Ontologies
GO
Topology
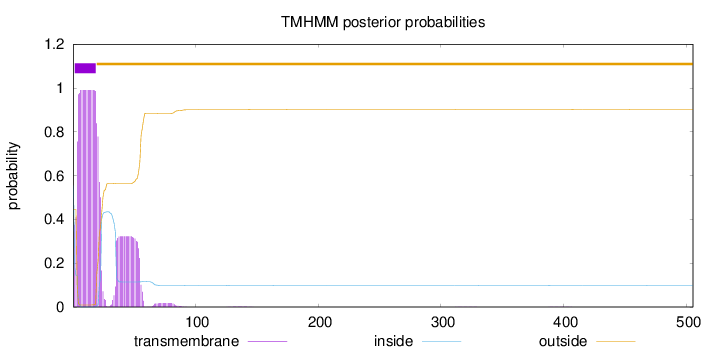
Length:
505
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
26.3288799999999
Exp number, first 60 AAs:
25.95969
Total prob of N-in:
0.55446
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 19
outside
20 - 505
Population Genetic Test Statistics
Pi
219.920354
Theta
24.942642
Tajima's D
1.851091
CLR
0.391394
CSRT
0.862556872156392
Interpretation
Uncertain
