Gene
KWMTBOMO02816 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003684
Annotation
PREDICTED:_glutaminase_kidney_isoform?_mitochondrial_isoform_X6_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.536
Sequence
CDS
ATGAACGCTAGCCAGCTCCGTCGACTCGCCTCACCGATTCTTAAGCCCTCGATATGCGCCTGTTTTCGACACTACAGTCAAGTGAGACCCGTCTATATAATTGAAAACTCTGATAAATGGCGCAACTCAGCACCCAACTTCTATTCGATTCAATCCGTCGATATAAGGCAGTATTCGTTCGTGCATAACATCGACCCTAAATCAGTCCGAGATGGTCCACATAAGAATGCTGATGATGTCCTATTTGATATGTTTAAGAATGAAGAAACAGGATTACTTCCAGTTGGAAAATTTTTGGCCGCCCTAAGAACCACCGGGATAAGAAAAAACGACGCACGTGTTAAAGAGGTAATGCATAGCCTGTACAAAGTTCACAAGGAATCTAATTTCGAAGGAGGATCTCCAGAAACACTGAAACTTGATCGAAAGACCTTCAAAGAAGTTATTGCACCAAATATAGTGTTGATAACTCGAGCTTTCCGAAGTCAGTTCGTAATTCCAGACTTCCAAGATTTTATCAAGGACATAGAGGAAATGTACTGGGCTGCAAAGAGCAATACTGATGGTAAAAATGCGAGCTATATCCCGCAATTAACGAGAGCCAGTCCCGAGAGTTGGGGCGTAAGTGTTTGCACAATTGATGGTCAACGTTTTTCTATTGGTGACGTGACCGTGCCATTTACTCTTCAATCTTGCAGCAAGCCTTTGACTTATGCAATGGCCCTTGAAACTCTTGGTCCTGACGTAGTGCACAACTATGTTGGCACAGAACCTAGCGGACGGAACTTTAATGAATTGATCCTCGATTATAATATGAAACCACACAACCCTATGATCAACGCTGGTGCTATTCTTGTTTGTTCATTACTAAAAACGCTGGACAAACCAGAAATGACATTGGCTGAAAAATTCGACTACGTCATGTCGTTCTTTAGTCGCCTTGCTGGGAATGAGGTGTTGGGCTTTAATAATGCAGTATTTTTATCGGAACGTGAAGCCGCCGACCGAAACTATGCTCTGGGTTTTTACATGAGGGAACACAAATGCTACCCCGAGAAAACAAATCTGAGGGAATGTATGGACTTTTATTTCCAGTGCTGTTCCATGGAAGCAAACTGCGATATAATGTCAATTATGGCGGCAACTTTGGCTAACGGAGGAATTTGTCCGATAACTGATGAAAAAGTATTGCGACCAGACTCCGTGCGTAATGTGCTGTCGTTAATGCACAGTTGCGGCATGTACGACTACTCTGGACAGTTCGCTTTCAAAGTGGGACTCCCTGCCAAATCTGGAGTATCGGGCGCCATGCTTATTGTGGTCCCAAATGTTATGGGCATATGTACATGGTCACCGCCTCTAGATCCATTGGGAAATAGCTGCAGAGGCTTACAGTTCTGCGATGACTTAATTAAACGATTCAATTTTCACAAATACGATAACATTCGTTACGCGAGCCACAAAAAGGATCCCCGCCGATACAAGTTCGAATCCACAGGACTTAATATCGTCAACTTATTGTTCTCTGCTGCAAGTGGAGATCTCAGTGCACTGAGAAGGCACCACCTGTCTGGTATGGACATGACTTTGTCGGACTACGATGGACGGACGGCTTTGCATTTATCCGCTGCTGAGGGTCATTTGGCGTGCGTCGAATTTTTGCTCGCTCAATGTGGAGTACCTCATGACCCGCGAGATCGTTGGGGCAGTCGACCTCTAAATGAAGCCGAAACATTTGGACACACTGCCGTTGTTCAATATCTGAAGGAATGGGAACGCGCTCACCCCGAAAACCCCCCAGTAGAATCACCATCCTCAGTGGACGAAATACTCGTGACTAAGCCAGATGCTACCGATGCTGTAAAGGATCCCAGCATCCAGGAGATAGTTACCAGAAATGGAGATAAAGTACAATAA
Protein
MNASQLRRLASPILKPSICACFRHYSQVRPVYIIENSDKWRNSAPNFYSIQSVDIRQYSFVHNIDPKSVRDGPHKNADDVLFDMFKNEETGLLPVGKFLAALRTTGIRKNDARVKEVMHSLYKVHKESNFEGGSPETLKLDRKTFKEVIAPNIVLITRAFRSQFVIPDFQDFIKDIEEMYWAAKSNTDGKNASYIPQLTRASPESWGVSVCTIDGQRFSIGDVTVPFTLQSCSKPLTYAMALETLGPDVVHNYVGTEPSGRNFNELILDYNMKPHNPMINAGAILVCSLLKTLDKPEMTLAEKFDYVMSFFSRLAGNEVLGFNNAVFLSEREAADRNYALGFYMREHKCYPEKTNLRECMDFYFQCCSMEANCDIMSIMAATLANGGICPITDEKVLRPDSVRNVLSLMHSCGMYDYSGQFAFKVGLPAKSGVSGAMLIVVPNVMGICTWSPPLDPLGNSCRGLQFCDDLIKRFNFHKYDNIRYASHKKDPRRYKFESTGLNIVNLLFSAASGDLSALRRHHLSGMDMTLSDYDGRTALHLSAAEGHLACVEFLLAQCGVPHDPRDRWGSRPLNEAETFGHTAVVQYLKEWERAHPENPPVESPSSVDEILVTKPDATDAVKDPSIQEIVTRNGDKVQ
Summary
Uniprot
A0A0G3VGT3
A0A194QT88
A0A194PUX6
A0A2A4JUS2
A0A212F1W0
A0A2W1BKW8
+ More
A0A2A4JTL2 A0A2H1V2C0 A0A2A4JV61 H9J2E7 A0A0L7LPD9 A0A2A4JVF2 A0A1B0CEM2 A0A232F046 K7ILT2 V5SK54 Q16YE5 A0A1S4FJT9 E2C8Y4 A0A1L8E2K5 A0A0C9QUJ2 V9IBK3 A0A1L8E2T3 A0A2A3EQL0 A0A158NUE0 A0A158NUE1 F4X7Y5 A0A0C9RBQ0 A0A1Q3FRJ4 A0A151WFY9 A0A1B6IUY0 A0A151INY3 A0A1B6G9X3 A0A182HBN0 A0A154PP86 A0A158NUE2 A0A1B6I2H0 A0A088ANW7 A0A310SIV9 A0A1B6J9P7 A0A195FXJ6 A0A151J124 A0A1B6H141 B0WH52 E1ZUU3 A0A1B6DQU4 A0A1B6DTV9 A0A195BSD5 V9IDX0 A0A1I8N988 T1PHJ5 A0A2M4CI44 A0A1I8N995 A0A2J7Q814 A0A1I8N992 A0A1I8N982 A0A067QW65 A0A2J7Q806 A0A182PCW4 A0A182F7W8 A0A0L0BNY2 A0A023F624 A0A182XWL4 A0A182VYA4 A0A1Y1N5E0 A0A336LR31 A0A182RFX7 A0A1I8N991 T1HK74 A0A182J0P9 A0A182M7V8 A0A2J7Q803 A0A2M4CP09 W5JRB2 A0A2M4CNV3 A0A1A9ZYR3 A0A182QTY2 A0A182XKB9 A0A182K786 A0A084WT45 A0A182V6T5 A0A182NSS9 A0A1J1ITI7 A0A182U636 A0A1I8QDE7 A0A182HJM3 A0A1B0GBY4 A0A1I8QDD6 A0A1A9V0S5 A0A1I8QDI2 A0A1Y1N515 A0A1I8QDC9 A0A0A1XM08 A0A1I8QDB3 A0A034VCD2 A0A034VHD4 A0A034VEM9 A0A1A9X3K8 A0A034VGP7 W8AEM7
A0A2A4JTL2 A0A2H1V2C0 A0A2A4JV61 H9J2E7 A0A0L7LPD9 A0A2A4JVF2 A0A1B0CEM2 A0A232F046 K7ILT2 V5SK54 Q16YE5 A0A1S4FJT9 E2C8Y4 A0A1L8E2K5 A0A0C9QUJ2 V9IBK3 A0A1L8E2T3 A0A2A3EQL0 A0A158NUE0 A0A158NUE1 F4X7Y5 A0A0C9RBQ0 A0A1Q3FRJ4 A0A151WFY9 A0A1B6IUY0 A0A151INY3 A0A1B6G9X3 A0A182HBN0 A0A154PP86 A0A158NUE2 A0A1B6I2H0 A0A088ANW7 A0A310SIV9 A0A1B6J9P7 A0A195FXJ6 A0A151J124 A0A1B6H141 B0WH52 E1ZUU3 A0A1B6DQU4 A0A1B6DTV9 A0A195BSD5 V9IDX0 A0A1I8N988 T1PHJ5 A0A2M4CI44 A0A1I8N995 A0A2J7Q814 A0A1I8N992 A0A1I8N982 A0A067QW65 A0A2J7Q806 A0A182PCW4 A0A182F7W8 A0A0L0BNY2 A0A023F624 A0A182XWL4 A0A182VYA4 A0A1Y1N5E0 A0A336LR31 A0A182RFX7 A0A1I8N991 T1HK74 A0A182J0P9 A0A182M7V8 A0A2J7Q803 A0A2M4CP09 W5JRB2 A0A2M4CNV3 A0A1A9ZYR3 A0A182QTY2 A0A182XKB9 A0A182K786 A0A084WT45 A0A182V6T5 A0A182NSS9 A0A1J1ITI7 A0A182U636 A0A1I8QDE7 A0A182HJM3 A0A1B0GBY4 A0A1I8QDD6 A0A1A9V0S5 A0A1I8QDI2 A0A1Y1N515 A0A1I8QDC9 A0A0A1XM08 A0A1I8QDB3 A0A034VCD2 A0A034VHD4 A0A034VEM9 A0A1A9X3K8 A0A034VGP7 W8AEM7
Pubmed
EMBL
KP657636
AKL78861.1
KQ461150
KPJ08738.1
KQ459597
KPI94935.1
+ More
NWSH01000604 PCG75374.1 AGBW02010786 OWR47719.1 KZ149992 PZC75538.1 PCG75375.1 ODYU01000355 SOQ34981.1 PCG75373.1 BABH01007783 JTDY01000393 KOB77393.1 PCG75372.1 AJWK01009000 NNAY01001459 OXU23900.1 KF647647 AHB50509.1 CH477517 EAT39659.1 GL453744 EFN75602.1 GFDF01001225 JAV12859.1 GBYB01004302 JAG74069.1 JR039062 AEY58513.1 GFDF01001202 JAV12882.1 KZ288193 PBC34055.1 ADTU01026390 GL888900 EGI57471.1 GBYB01004306 JAG74073.1 GFDL01004887 JAV30158.1 KQ983203 KYQ46741.1 GECU01019012 GECU01016960 GECU01007087 GECU01003230 JAS88694.1 JAS90746.1 JAT00620.1 JAT04477.1 KQ976899 KYN07179.1 GECZ01030363 GECZ01017153 GECZ01010676 GECZ01007974 GECZ01006277 GECZ01002385 GECZ01001988 JAS39406.1 JAS52616.1 JAS59093.1 JAS61795.1 JAS63492.1 JAS67384.1 JAS67781.1 JXUM01125345 JXUM01125346 KQ566934 KXJ69720.1 KQ435007 KZC13672.1 GECU01026580 GECU01014939 JAS81126.1 JAS92767.1 KQ762207 OAD56020.1 GECU01017608 GECU01011883 JAS90098.1 JAS95823.1 KQ981193 KYN45158.1 KQ980575 KYN15520.1 GECZ01001363 JAS68406.1 DS231933 EDS27559.1 GL434297 EFN75035.1 GEDC01009242 GEDC01000519 JAS28056.1 JAS36779.1 GEDC01008184 JAS29114.1 KQ976417 KYM89930.1 JR039063 AEY58514.1 KA647363 AFP61992.1 GGFL01000741 MBW64919.1 NEVH01016978 PNF24715.1 KK852886 KDR14351.1 PNF24717.1 JRES01001578 KNC21795.1 GBBI01002055 JAC16657.1 GEZM01012350 GEZM01012347 GEZM01012346 JAV92979.1 UFQS01000126 UFQT01000126 SSX00132.1 SSX20512.1 ACPB03011909 AXCM01006045 PNF24716.1 GGFL01002827 MBW67005.1 ADMH02000406 ETN66676.1 GGFL01002826 MBW67004.1 AXCN02000300 ATLV01026836 KE525419 KFB53389.1 CVRI01000059 CRL03549.1 APCN01002188 CCAG010005902 GEZM01012345 JAV92982.1 GBXI01006894 GBXI01002402 JAD07398.1 JAD11890.1 GAKP01017981 JAC40971.1 GAKP01017979 JAC40973.1 GAKP01017978 JAC40974.1 GAKP01017982 JAC40970.1 GAMC01019896 GAMC01019895 JAB86660.1
NWSH01000604 PCG75374.1 AGBW02010786 OWR47719.1 KZ149992 PZC75538.1 PCG75375.1 ODYU01000355 SOQ34981.1 PCG75373.1 BABH01007783 JTDY01000393 KOB77393.1 PCG75372.1 AJWK01009000 NNAY01001459 OXU23900.1 KF647647 AHB50509.1 CH477517 EAT39659.1 GL453744 EFN75602.1 GFDF01001225 JAV12859.1 GBYB01004302 JAG74069.1 JR039062 AEY58513.1 GFDF01001202 JAV12882.1 KZ288193 PBC34055.1 ADTU01026390 GL888900 EGI57471.1 GBYB01004306 JAG74073.1 GFDL01004887 JAV30158.1 KQ983203 KYQ46741.1 GECU01019012 GECU01016960 GECU01007087 GECU01003230 JAS88694.1 JAS90746.1 JAT00620.1 JAT04477.1 KQ976899 KYN07179.1 GECZ01030363 GECZ01017153 GECZ01010676 GECZ01007974 GECZ01006277 GECZ01002385 GECZ01001988 JAS39406.1 JAS52616.1 JAS59093.1 JAS61795.1 JAS63492.1 JAS67384.1 JAS67781.1 JXUM01125345 JXUM01125346 KQ566934 KXJ69720.1 KQ435007 KZC13672.1 GECU01026580 GECU01014939 JAS81126.1 JAS92767.1 KQ762207 OAD56020.1 GECU01017608 GECU01011883 JAS90098.1 JAS95823.1 KQ981193 KYN45158.1 KQ980575 KYN15520.1 GECZ01001363 JAS68406.1 DS231933 EDS27559.1 GL434297 EFN75035.1 GEDC01009242 GEDC01000519 JAS28056.1 JAS36779.1 GEDC01008184 JAS29114.1 KQ976417 KYM89930.1 JR039063 AEY58514.1 KA647363 AFP61992.1 GGFL01000741 MBW64919.1 NEVH01016978 PNF24715.1 KK852886 KDR14351.1 PNF24717.1 JRES01001578 KNC21795.1 GBBI01002055 JAC16657.1 GEZM01012350 GEZM01012347 GEZM01012346 JAV92979.1 UFQS01000126 UFQT01000126 SSX00132.1 SSX20512.1 ACPB03011909 AXCM01006045 PNF24716.1 GGFL01002827 MBW67005.1 ADMH02000406 ETN66676.1 GGFL01002826 MBW67004.1 AXCN02000300 ATLV01026836 KE525419 KFB53389.1 CVRI01000059 CRL03549.1 APCN01002188 CCAG010005902 GEZM01012345 JAV92982.1 GBXI01006894 GBXI01002402 JAD07398.1 JAD11890.1 GAKP01017981 JAC40971.1 GAKP01017979 JAC40973.1 GAKP01017978 JAC40974.1 GAKP01017982 JAC40970.1 GAMC01019896 GAMC01019895 JAB86660.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007151
UP000005204
UP000037510
+ More
UP000092461 UP000215335 UP000002358 UP000008820 UP000008237 UP000242457 UP000005205 UP000007755 UP000075809 UP000078542 UP000069940 UP000249989 UP000076502 UP000005203 UP000078541 UP000078492 UP000002320 UP000000311 UP000078540 UP000095301 UP000235965 UP000027135 UP000075885 UP000069272 UP000037069 UP000076408 UP000075920 UP000075900 UP000015103 UP000075880 UP000075883 UP000000673 UP000092445 UP000075886 UP000076407 UP000075881 UP000030765 UP000075903 UP000075884 UP000183832 UP000075902 UP000095300 UP000075840 UP000092444 UP000078200 UP000091820
UP000092461 UP000215335 UP000002358 UP000008820 UP000008237 UP000242457 UP000005205 UP000007755 UP000075809 UP000078542 UP000069940 UP000249989 UP000076502 UP000005203 UP000078541 UP000078492 UP000002320 UP000000311 UP000078540 UP000095301 UP000235965 UP000027135 UP000075885 UP000069272 UP000037069 UP000076408 UP000075920 UP000075900 UP000015103 UP000075880 UP000075883 UP000000673 UP000092445 UP000075886 UP000076407 UP000075881 UP000030765 UP000075903 UP000075884 UP000183832 UP000075902 UP000095300 UP000075840 UP000092444 UP000078200 UP000091820
PRIDE
Pfam
Interpro
IPR036770
Ankyrin_rpt-contain_sf
+ More
IPR020683 Ankyrin_rpt-contain_dom
IPR015868 Glutaminase
IPR012338 Beta-lactam/transpept-like
IPR041541 Glutaminase_EF-hand
IPR002110 Ankyrin_rpt
IPR005120 UPF3_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035979 RBD_domain_sf
IPR019375 Ribosomal_S28_mit
IPR008516 Na/K-Atpase_Interacting
IPR004942 Roadblock/LAMTOR2_dom
IPR006593 Cyt_b561/ferric_Rdtase_TM
IPR020683 Ankyrin_rpt-contain_dom
IPR015868 Glutaminase
IPR012338 Beta-lactam/transpept-like
IPR041541 Glutaminase_EF-hand
IPR002110 Ankyrin_rpt
IPR005120 UPF3_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR035979 RBD_domain_sf
IPR019375 Ribosomal_S28_mit
IPR008516 Na/K-Atpase_Interacting
IPR004942 Roadblock/LAMTOR2_dom
IPR006593 Cyt_b561/ferric_Rdtase_TM
Gene 3D
CDD
ProteinModelPortal
A0A0G3VGT3
A0A194QT88
A0A194PUX6
A0A2A4JUS2
A0A212F1W0
A0A2W1BKW8
+ More
A0A2A4JTL2 A0A2H1V2C0 A0A2A4JV61 H9J2E7 A0A0L7LPD9 A0A2A4JVF2 A0A1B0CEM2 A0A232F046 K7ILT2 V5SK54 Q16YE5 A0A1S4FJT9 E2C8Y4 A0A1L8E2K5 A0A0C9QUJ2 V9IBK3 A0A1L8E2T3 A0A2A3EQL0 A0A158NUE0 A0A158NUE1 F4X7Y5 A0A0C9RBQ0 A0A1Q3FRJ4 A0A151WFY9 A0A1B6IUY0 A0A151INY3 A0A1B6G9X3 A0A182HBN0 A0A154PP86 A0A158NUE2 A0A1B6I2H0 A0A088ANW7 A0A310SIV9 A0A1B6J9P7 A0A195FXJ6 A0A151J124 A0A1B6H141 B0WH52 E1ZUU3 A0A1B6DQU4 A0A1B6DTV9 A0A195BSD5 V9IDX0 A0A1I8N988 T1PHJ5 A0A2M4CI44 A0A1I8N995 A0A2J7Q814 A0A1I8N992 A0A1I8N982 A0A067QW65 A0A2J7Q806 A0A182PCW4 A0A182F7W8 A0A0L0BNY2 A0A023F624 A0A182XWL4 A0A182VYA4 A0A1Y1N5E0 A0A336LR31 A0A182RFX7 A0A1I8N991 T1HK74 A0A182J0P9 A0A182M7V8 A0A2J7Q803 A0A2M4CP09 W5JRB2 A0A2M4CNV3 A0A1A9ZYR3 A0A182QTY2 A0A182XKB9 A0A182K786 A0A084WT45 A0A182V6T5 A0A182NSS9 A0A1J1ITI7 A0A182U636 A0A1I8QDE7 A0A182HJM3 A0A1B0GBY4 A0A1I8QDD6 A0A1A9V0S5 A0A1I8QDI2 A0A1Y1N515 A0A1I8QDC9 A0A0A1XM08 A0A1I8QDB3 A0A034VCD2 A0A034VHD4 A0A034VEM9 A0A1A9X3K8 A0A034VGP7 W8AEM7
A0A2A4JTL2 A0A2H1V2C0 A0A2A4JV61 H9J2E7 A0A0L7LPD9 A0A2A4JVF2 A0A1B0CEM2 A0A232F046 K7ILT2 V5SK54 Q16YE5 A0A1S4FJT9 E2C8Y4 A0A1L8E2K5 A0A0C9QUJ2 V9IBK3 A0A1L8E2T3 A0A2A3EQL0 A0A158NUE0 A0A158NUE1 F4X7Y5 A0A0C9RBQ0 A0A1Q3FRJ4 A0A151WFY9 A0A1B6IUY0 A0A151INY3 A0A1B6G9X3 A0A182HBN0 A0A154PP86 A0A158NUE2 A0A1B6I2H0 A0A088ANW7 A0A310SIV9 A0A1B6J9P7 A0A195FXJ6 A0A151J124 A0A1B6H141 B0WH52 E1ZUU3 A0A1B6DQU4 A0A1B6DTV9 A0A195BSD5 V9IDX0 A0A1I8N988 T1PHJ5 A0A2M4CI44 A0A1I8N995 A0A2J7Q814 A0A1I8N992 A0A1I8N982 A0A067QW65 A0A2J7Q806 A0A182PCW4 A0A182F7W8 A0A0L0BNY2 A0A023F624 A0A182XWL4 A0A182VYA4 A0A1Y1N5E0 A0A336LR31 A0A182RFX7 A0A1I8N991 T1HK74 A0A182J0P9 A0A182M7V8 A0A2J7Q803 A0A2M4CP09 W5JRB2 A0A2M4CNV3 A0A1A9ZYR3 A0A182QTY2 A0A182XKB9 A0A182K786 A0A084WT45 A0A182V6T5 A0A182NSS9 A0A1J1ITI7 A0A182U636 A0A1I8QDE7 A0A182HJM3 A0A1B0GBY4 A0A1I8QDD6 A0A1A9V0S5 A0A1I8QDI2 A0A1Y1N515 A0A1I8QDC9 A0A0A1XM08 A0A1I8QDB3 A0A034VCD2 A0A034VHD4 A0A034VEM9 A0A1A9X3K8 A0A034VGP7 W8AEM7
PDB
5UQE
E-value=2.18756e-171,
Score=1548
Ontologies
PATHWAY
GO
PANTHER
Topology
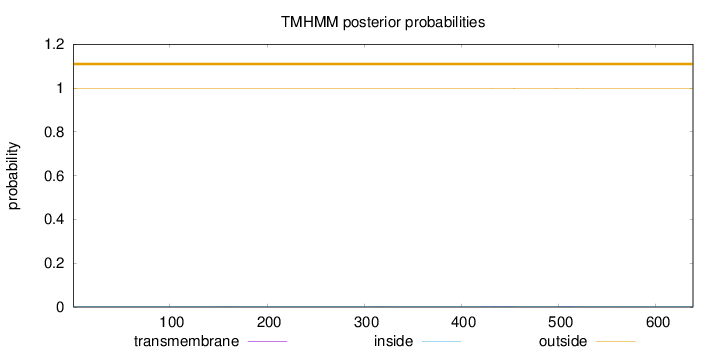
Length:
638
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0850399999999999
Exp number, first 60 AAs:
0.00037
Total prob of N-in:
0.00217
outside
1 - 638
Population Genetic Test Statistics
Pi
261.351583
Theta
223.621788
Tajima's D
0.990882
CLR
0.000892
CSRT
0.660816959152042
Interpretation
Uncertain
