Gene
KWMTBOMO02815 Validated by peptides from experiments
Annotation
Mitochondrial_28S_ribosomal_protein_S28_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.195 Nuclear Reliability : 1.554
Sequence
CDS
ATGGATTTGGGGGATCCAGTGGGAAAAACCGTAATAGGAAAAATATTTCATGTTGTAGAAGACGACTTATATATAGATTTTGGATGGAAATTTCATTGTGTTTGTACAAGACCAAATTTGAGAGGTGAAGAATACGTCAGGGGCACTAGAGTTCGGATACAAATTAATGATTTAGAATTATCATCTCGTTTCTTAGGATCGAGCACAGATCTTACACTTTTAGAGGCAGACTGTAAATTACTTGGTATTATATCTACTCCCACTAAACCAGCTGTAAGTGTTAATTAA
Protein
MDLGDPVGKTVIGKIFHVVEDDLYIDFGWKFHCVCTRPNLRGEEYVRGTRVRIQINDLELSSRFLGSSTDLTLLEADCKLLGIISTPTKPAVSVN
Summary
Uniprot
A0A0L7LPR1
A0A2H1V2B3
A0A194PQB8
A0A212F1V2
A0A2A4JTN3
A0A1E1WTJ2
+ More
A0A1E1WM93 B0WEG4 A0A2M4AR20 A0A2M4ARI9 A0A2M4AR35 A0A2M4C0E3 A0A182K1S8 A0A2M4C0J4 A0A2M4C0G1 A0A2M4C0T4 A0A2M3ZKL1 W5JB38 A0A2M3ZC03 A0A182FDB6 A0A182MKK3 A0A2M3ZJF8 A0A023EJ77 Q7Q544 A0A182X4N0 A0A182TWA5 A0A182LK89 A0A182I4M8 A0A182G4S7 A0A1L8E0C7 A0A182VEP8 A0A1Q3FBR2 A0A182WBM0 A0A182QID6 A0A182RDK2 A0A182PDJ9 A0A1A9ZXI1 Q28ZM9 A0A1I8MLK2 B4GH17 A0A182NI60 A0A1L8E060 B4KSU1 A0A0L0BMN8 A0A1B0C3R8 B4LM24 W8BR70 A0A1A9XPI7 A0A034WWW8 A0A3B0JPM1 A0A0K8UG64 A0A1A9X4S2 A0A1I8Q9W5 Q17EN8 A0A1A9V256 Q6XHU9 A0A1B6GDH8 B3NMU9 A0A1B6HU85 A0A2P8ZNF1 A0A0M4EGX5 A0A1B0D663 B4P4C4 A0A1W4VPS0 A0A2J7QJ93 A0A158NB29 A0A151I1X7 B4QCZ6 B3MDF3 F4W3U1 B4J524 E9IZX7 A0A1B6MTC5 A0A151IQB1 F4WCW5 A0A0J7KA33 A0A026X192 A1ZBA5 A0A3L8DL47 A0A1W4WTA4 A0A151WZS3 B4MRC3 N6T0A9 A0A195DZI6 E2A4L1 J3JW93 A0A1Y1KWY7 A0A067QHQ0 A0A0N7Z8F2 Q8SX15 A0A195EWN7 T1HTI9 A0A146MCZ8 A0A0K8R9B8 A0A0K8SFQ7 A0A0A9VYH0 A0A2R7WBL2 E2C6B5 A0A1B6DPT8 A0A0C9RHD1
A0A1E1WM93 B0WEG4 A0A2M4AR20 A0A2M4ARI9 A0A2M4AR35 A0A2M4C0E3 A0A182K1S8 A0A2M4C0J4 A0A2M4C0G1 A0A2M4C0T4 A0A2M3ZKL1 W5JB38 A0A2M3ZC03 A0A182FDB6 A0A182MKK3 A0A2M3ZJF8 A0A023EJ77 Q7Q544 A0A182X4N0 A0A182TWA5 A0A182LK89 A0A182I4M8 A0A182G4S7 A0A1L8E0C7 A0A182VEP8 A0A1Q3FBR2 A0A182WBM0 A0A182QID6 A0A182RDK2 A0A182PDJ9 A0A1A9ZXI1 Q28ZM9 A0A1I8MLK2 B4GH17 A0A182NI60 A0A1L8E060 B4KSU1 A0A0L0BMN8 A0A1B0C3R8 B4LM24 W8BR70 A0A1A9XPI7 A0A034WWW8 A0A3B0JPM1 A0A0K8UG64 A0A1A9X4S2 A0A1I8Q9W5 Q17EN8 A0A1A9V256 Q6XHU9 A0A1B6GDH8 B3NMU9 A0A1B6HU85 A0A2P8ZNF1 A0A0M4EGX5 A0A1B0D663 B4P4C4 A0A1W4VPS0 A0A2J7QJ93 A0A158NB29 A0A151I1X7 B4QCZ6 B3MDF3 F4W3U1 B4J524 E9IZX7 A0A1B6MTC5 A0A151IQB1 F4WCW5 A0A0J7KA33 A0A026X192 A1ZBA5 A0A3L8DL47 A0A1W4WTA4 A0A151WZS3 B4MRC3 N6T0A9 A0A195DZI6 E2A4L1 J3JW93 A0A1Y1KWY7 A0A067QHQ0 A0A0N7Z8F2 Q8SX15 A0A195EWN7 T1HTI9 A0A146MCZ8 A0A0K8R9B8 A0A0K8SFQ7 A0A0A9VYH0 A0A2R7WBL2 E2C6B5 A0A1B6DPT8 A0A0C9RHD1
Pubmed
26227816
26354079
22118469
20920257
23761445
24945155
+ More
12364791 14747013 17210077 20966253 26483478 15632085 17994087 25315136 18057021 26108605 24495485 25348373 17510324 14525923 29403074 17550304 21347285 22936249 21719571 21282665 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 23537049 20798317 22516182 28004739 24845553 27129103 26823975 25401762
12364791 14747013 17210077 20966253 26483478 15632085 17994087 25315136 18057021 26108605 24495485 25348373 17510324 14525923 29403074 17550304 21347285 22936249 21719571 21282665 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 23537049 20798317 22516182 28004739 24845553 27129103 26823975 25401762
EMBL
JTDY01000393
KOB77389.1
ODYU01000355
SOQ34980.1
KQ459597
KPI94934.1
+ More
AGBW02010786 OWR47718.1 NWSH01000604 PCG75371.1 GDQN01000898 JAT90156.1 GDQN01002986 JAT88068.1 DS231909 EDS45624.1 GGFK01009915 MBW43236.1 GGFK01009897 MBW43218.1 GGFK01009861 MBW43182.1 GGFJ01009649 MBW58790.1 GGFJ01009651 MBW58792.1 GGFJ01009652 MBW58793.1 GGFJ01009650 MBW58791.1 GGFM01008284 MBW29035.1 ADMH02001892 ETN60623.1 GGFM01005227 MBW25978.1 AXCM01003043 GGFM01007966 MBW28717.1 GAPW01004613 JAC08985.1 AAAB01008960 EAA10773.3 APCN01002331 JXUM01042784 KQ561338 KXJ78921.1 GFDF01001906 JAV12178.1 GFDL01010058 JAV24987.1 AXCN02000395 CM000071 EAL25584.1 CH479183 EDW35787.1 GFDF01001936 JAV12148.1 CH933808 EDW08438.1 KRG04129.1 JRES01001648 KNC21198.1 JXJN01025096 CH940648 EDW59944.1 GAMC01007227 JAB99328.1 GAKP01000679 JAC58273.1 OUUW01000001 SPP75579.1 GDHF01026647 JAI25667.1 CH477280 EAT44955.1 AY232084 AAR10107.1 GECZ01009298 JAS60471.1 CH954179 EDV55103.1 GECU01029469 JAS78237.1 PYGN01000010 PSN58041.1 CP012524 ALC41735.1 AJVK01025773 CM000158 EDW91610.1 NEVH01013559 PNF28661.1 ADTU01010783 KQ976572 KYM80410.1 CM000362 CM002911 EDX07701.1 KMY94886.1 CH902619 EDV36401.1 GL887446 EGI71132.1 CH916367 EDW01730.1 GL767291 EFZ13822.1 GEBQ01000832 JAT39145.1 KQ976782 KYN08310.1 GL888074 EGI67946.1 LBMM01010899 KMQ87167.1 KK107039 EZA61863.1 AE013599 BT044321 AAF57671.2 ACH92386.1 QOIP01000006 RLU21165.1 KQ982630 KYQ53398.1 CH963850 EDW74662.2 APGK01057485 KB741280 KB632033 ENN70943.1 ERL88185.1 KQ980050 KYN18039.1 GL436710 EFN71613.1 BT127511 AEE62473.1 GEZM01071357 GEZM01071356 JAV65872.1 KK853606 KDR06171.1 GDKW01003515 JAI53080.1 AY094907 AAM11260.1 KQ981953 KYN32312.1 ACPB03017753 GDHC01002013 JAQ16616.1 GADI01006152 JAA67656.1 GBRD01014226 JAG51600.1 GBHO01043373 JAG00231.1 KK854582 PTY17084.1 GL453058 EFN76514.1 GEDC01009593 JAS27705.1 GBYB01012612 JAG82379.1
AGBW02010786 OWR47718.1 NWSH01000604 PCG75371.1 GDQN01000898 JAT90156.1 GDQN01002986 JAT88068.1 DS231909 EDS45624.1 GGFK01009915 MBW43236.1 GGFK01009897 MBW43218.1 GGFK01009861 MBW43182.1 GGFJ01009649 MBW58790.1 GGFJ01009651 MBW58792.1 GGFJ01009652 MBW58793.1 GGFJ01009650 MBW58791.1 GGFM01008284 MBW29035.1 ADMH02001892 ETN60623.1 GGFM01005227 MBW25978.1 AXCM01003043 GGFM01007966 MBW28717.1 GAPW01004613 JAC08985.1 AAAB01008960 EAA10773.3 APCN01002331 JXUM01042784 KQ561338 KXJ78921.1 GFDF01001906 JAV12178.1 GFDL01010058 JAV24987.1 AXCN02000395 CM000071 EAL25584.1 CH479183 EDW35787.1 GFDF01001936 JAV12148.1 CH933808 EDW08438.1 KRG04129.1 JRES01001648 KNC21198.1 JXJN01025096 CH940648 EDW59944.1 GAMC01007227 JAB99328.1 GAKP01000679 JAC58273.1 OUUW01000001 SPP75579.1 GDHF01026647 JAI25667.1 CH477280 EAT44955.1 AY232084 AAR10107.1 GECZ01009298 JAS60471.1 CH954179 EDV55103.1 GECU01029469 JAS78237.1 PYGN01000010 PSN58041.1 CP012524 ALC41735.1 AJVK01025773 CM000158 EDW91610.1 NEVH01013559 PNF28661.1 ADTU01010783 KQ976572 KYM80410.1 CM000362 CM002911 EDX07701.1 KMY94886.1 CH902619 EDV36401.1 GL887446 EGI71132.1 CH916367 EDW01730.1 GL767291 EFZ13822.1 GEBQ01000832 JAT39145.1 KQ976782 KYN08310.1 GL888074 EGI67946.1 LBMM01010899 KMQ87167.1 KK107039 EZA61863.1 AE013599 BT044321 AAF57671.2 ACH92386.1 QOIP01000006 RLU21165.1 KQ982630 KYQ53398.1 CH963850 EDW74662.2 APGK01057485 KB741280 KB632033 ENN70943.1 ERL88185.1 KQ980050 KYN18039.1 GL436710 EFN71613.1 BT127511 AEE62473.1 GEZM01071357 GEZM01071356 JAV65872.1 KK853606 KDR06171.1 GDKW01003515 JAI53080.1 AY094907 AAM11260.1 KQ981953 KYN32312.1 ACPB03017753 GDHC01002013 JAQ16616.1 GADI01006152 JAA67656.1 GBRD01014226 JAG51600.1 GBHO01043373 JAG00231.1 KK854582 PTY17084.1 GL453058 EFN76514.1 GEDC01009593 JAS27705.1 GBYB01012612 JAG82379.1
Proteomes
UP000037510
UP000053268
UP000007151
UP000218220
UP000002320
UP000075881
+ More
UP000000673 UP000069272 UP000075883 UP000007062 UP000076407 UP000075902 UP000075882 UP000075840 UP000069940 UP000249989 UP000075903 UP000075920 UP000075886 UP000075900 UP000075885 UP000092445 UP000001819 UP000095301 UP000008744 UP000075884 UP000009192 UP000037069 UP000092460 UP000008792 UP000092443 UP000268350 UP000091820 UP000095300 UP000008820 UP000078200 UP000008711 UP000245037 UP000092553 UP000092462 UP000002282 UP000192221 UP000235965 UP000005205 UP000078540 UP000000304 UP000007801 UP000007755 UP000001070 UP000078542 UP000036403 UP000053097 UP000000803 UP000279307 UP000192223 UP000075809 UP000007798 UP000019118 UP000030742 UP000078492 UP000000311 UP000027135 UP000078541 UP000015103 UP000008237
UP000000673 UP000069272 UP000075883 UP000007062 UP000076407 UP000075902 UP000075882 UP000075840 UP000069940 UP000249989 UP000075903 UP000075920 UP000075886 UP000075900 UP000075885 UP000092445 UP000001819 UP000095301 UP000008744 UP000075884 UP000009192 UP000037069 UP000092460 UP000008792 UP000092443 UP000268350 UP000091820 UP000095300 UP000008820 UP000078200 UP000008711 UP000245037 UP000092553 UP000092462 UP000002282 UP000192221 UP000235965 UP000005205 UP000078540 UP000000304 UP000007801 UP000007755 UP000001070 UP000078542 UP000036403 UP000053097 UP000000803 UP000279307 UP000192223 UP000075809 UP000007798 UP000019118 UP000030742 UP000078492 UP000000311 UP000027135 UP000078541 UP000015103 UP000008237
Pfam
PF10246 MRP-S35
Interpro
IPR019375
Ribosomal_S28_mit
ProteinModelPortal
A0A0L7LPR1
A0A2H1V2B3
A0A194PQB8
A0A212F1V2
A0A2A4JTN3
A0A1E1WTJ2
+ More
A0A1E1WM93 B0WEG4 A0A2M4AR20 A0A2M4ARI9 A0A2M4AR35 A0A2M4C0E3 A0A182K1S8 A0A2M4C0J4 A0A2M4C0G1 A0A2M4C0T4 A0A2M3ZKL1 W5JB38 A0A2M3ZC03 A0A182FDB6 A0A182MKK3 A0A2M3ZJF8 A0A023EJ77 Q7Q544 A0A182X4N0 A0A182TWA5 A0A182LK89 A0A182I4M8 A0A182G4S7 A0A1L8E0C7 A0A182VEP8 A0A1Q3FBR2 A0A182WBM0 A0A182QID6 A0A182RDK2 A0A182PDJ9 A0A1A9ZXI1 Q28ZM9 A0A1I8MLK2 B4GH17 A0A182NI60 A0A1L8E060 B4KSU1 A0A0L0BMN8 A0A1B0C3R8 B4LM24 W8BR70 A0A1A9XPI7 A0A034WWW8 A0A3B0JPM1 A0A0K8UG64 A0A1A9X4S2 A0A1I8Q9W5 Q17EN8 A0A1A9V256 Q6XHU9 A0A1B6GDH8 B3NMU9 A0A1B6HU85 A0A2P8ZNF1 A0A0M4EGX5 A0A1B0D663 B4P4C4 A0A1W4VPS0 A0A2J7QJ93 A0A158NB29 A0A151I1X7 B4QCZ6 B3MDF3 F4W3U1 B4J524 E9IZX7 A0A1B6MTC5 A0A151IQB1 F4WCW5 A0A0J7KA33 A0A026X192 A1ZBA5 A0A3L8DL47 A0A1W4WTA4 A0A151WZS3 B4MRC3 N6T0A9 A0A195DZI6 E2A4L1 J3JW93 A0A1Y1KWY7 A0A067QHQ0 A0A0N7Z8F2 Q8SX15 A0A195EWN7 T1HTI9 A0A146MCZ8 A0A0K8R9B8 A0A0K8SFQ7 A0A0A9VYH0 A0A2R7WBL2 E2C6B5 A0A1B6DPT8 A0A0C9RHD1
A0A1E1WM93 B0WEG4 A0A2M4AR20 A0A2M4ARI9 A0A2M4AR35 A0A2M4C0E3 A0A182K1S8 A0A2M4C0J4 A0A2M4C0G1 A0A2M4C0T4 A0A2M3ZKL1 W5JB38 A0A2M3ZC03 A0A182FDB6 A0A182MKK3 A0A2M3ZJF8 A0A023EJ77 Q7Q544 A0A182X4N0 A0A182TWA5 A0A182LK89 A0A182I4M8 A0A182G4S7 A0A1L8E0C7 A0A182VEP8 A0A1Q3FBR2 A0A182WBM0 A0A182QID6 A0A182RDK2 A0A182PDJ9 A0A1A9ZXI1 Q28ZM9 A0A1I8MLK2 B4GH17 A0A182NI60 A0A1L8E060 B4KSU1 A0A0L0BMN8 A0A1B0C3R8 B4LM24 W8BR70 A0A1A9XPI7 A0A034WWW8 A0A3B0JPM1 A0A0K8UG64 A0A1A9X4S2 A0A1I8Q9W5 Q17EN8 A0A1A9V256 Q6XHU9 A0A1B6GDH8 B3NMU9 A0A1B6HU85 A0A2P8ZNF1 A0A0M4EGX5 A0A1B0D663 B4P4C4 A0A1W4VPS0 A0A2J7QJ93 A0A158NB29 A0A151I1X7 B4QCZ6 B3MDF3 F4W3U1 B4J524 E9IZX7 A0A1B6MTC5 A0A151IQB1 F4WCW5 A0A0J7KA33 A0A026X192 A1ZBA5 A0A3L8DL47 A0A1W4WTA4 A0A151WZS3 B4MRC3 N6T0A9 A0A195DZI6 E2A4L1 J3JW93 A0A1Y1KWY7 A0A067QHQ0 A0A0N7Z8F2 Q8SX15 A0A195EWN7 T1HTI9 A0A146MCZ8 A0A0K8R9B8 A0A0K8SFQ7 A0A0A9VYH0 A0A2R7WBL2 E2C6B5 A0A1B6DPT8 A0A0C9RHD1
PDB
6GAZ
E-value=6.92439e-28,
Score=301
Ontologies
PANTHER
Topology
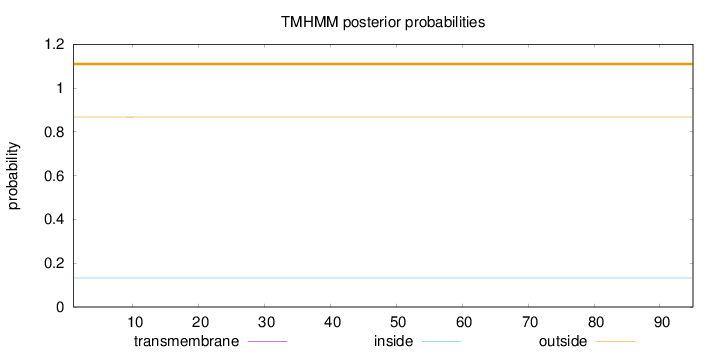
Length:
95
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00163
Exp number, first 60 AAs:
0.00114
Total prob of N-in:
0.13258
outside
1 - 95
Population Genetic Test Statistics
Pi
182.366804
Theta
154.379281
Tajima's D
0.701402
CLR
0
CSRT
0.574121293935303
Interpretation
Uncertain
