Gene
KWMTBOMO02812
Pre Gene Modal
BGIBMGA003686
Annotation
MSL2_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.544
Sequence
CDS
ATGAATGCAACAAGCCTTTACGTTTCTACGTGTCGACTCATAATATTAGCTGATCCCGCTGACAAATCTACGTGGACTGATTTATTTCGTTTGGTTCCGTATTTAAGACAATCTTTATCCTGTACGGTTTGCGGTAATTTACTAAAAGAACCATATACGCCAACAAGTTCTGGGTGCCAACATCACGTGTGTAAAAAGTGTAAAGGTGGTCGAAAAAAATTAAAACCTTCATGCAGTTGGTGTAAAGATTATGAAAATTATACTGAAAACTTACAGTTACGTATACTCCTTCAGTGTTACAAAAAGTTGTGCGAATATTTCATGGGAACAGAAGTGTACAAAACATTATTAGATGAAGATGAAGTTGCTGCCAGTGTGAATGGGGGTACAGTGGCCAGCTCAGGCTTAATAGATTTAATTCAGGAAGGTGCGGGATTTTCAGACGACTATAAAAGTACAGGAGGCCTTTCGAAGTCAGCATATAGCATATTACCATGTGTATACACAAATTCAGCATCAACACAAACCCAACATGGATCTAGTTCAAGTAGCTCGGGAGCAAGAAGTTCCAGAGATTCTCCCAATTCAAGATCAACAGCAAACGGATCTCCCATGTATTCTGTAATGTATGCTGGCTCTGGGAATAAAATCACAATCAAAAGAAAAGCAATGGATGAATTGGATTCACCCCAAAGTGAATCATCATCTAGGGAGAGTAAGTCACTCGGTTTCAAGAAACCATCAAATAGATCCCGTAATTCTACGGGCGGTAAGCGCAAAGGCTGCAGATGTGGAAATGCAACGGCAACACCGGGGAAACTTACTTGTTGCGGCCAAAGATGCCCCTGTTACGTTGACAGTAAACCTTGTACCGAGTGTAAGTGTAAAGGATGCAGAAATCCTCACAGGCCAGACGGGATGAAGGTCCGACCTCACATACCACAATTAGTGGACACACTGCAATTGACTCTAAACAACCCTGACAGCTCACCATCATGTTCAGGCTCAATGGACAGTTTGGATACTGATACCCTTACCATGGAGGCTATGGAGGAATCGCTTAGTTACACTGCAGAACTTAAATCGTCAAACATTAAAGTTTACACAAGTCAATTACAAGAGGTATCACCGTCGTTGCCAGCGACGATAATGATGGACGAGGACCTAGCTGAATCGCCGGACGATGATATGAGCTCCCCACATGACTACACCCTGGCATCTCCTCAGGGTTACGACGTGGACGTCATCTCACCGCAGTCAGCAGAGACTCAAGATGTCACCAGTGACATTGAGGTGGATTAG
Protein
MNATSLYVSTCRLIILADPADKSTWTDLFRLVPYLRQSLSCTVCGNLLKEPYTPTSSGCQHHVCKKCKGGRKKLKPSCSWCKDYENYTENLQLRILLQCYKKLCEYFMGTEVYKTLLDEDEVAASVNGGTVASSGLIDLIQEGAGFSDDYKSTGGLSKSAYSILPCVYTNSASTQTQHGSSSSSSGARSSRDSPNSRSTANGSPMYSVMYAGSGNKITIKRKAMDELDSPQSESSSRESKSLGFKKPSNRSRNSTGGKRKGCRCGNATATPGKLTCCGQRCPCYVDSKPCTECKCKGCRNPHRPDGMKVRPHIPQLVDTLQLTLNNPDSSPSCSGSMDSLDTDTLTMEAMEESLSYTAELKSSNIKVYTSQLQEVSPSLPATIMMDEDLAESPDDDMSSPHDYTLASPQGYDVDVISPQSAETQDVTSDIEVD
Summary
Uniprot
A5JPL8
A0A2W1BJG9
A0A2A4JUJ6
A0A2H1V2A6
A0A212F1U9
A0A194QT92
+ More
A0A194PNC0 A0A067QWJ4 E2C5R7 A0A088AVX8 E9IUQ8 A0A310SVH7 F4WZP1 E2ACB4 A0A026X1C9 A0A3L8DRC8 K7IQY9 A0A2A3E2B1 A0A0L7QYB7 A0A195B7S1 A0A158NGT5 A0A195FFF8 A0A151XCW6 A0A195DXT7 A0A232F1V5 A0A1L8DSQ6 A0A154PR83 A0A1B0CDV0 D6W8B3 A0A1B6EQT1 A0A1Y1KNL2 A0A1B0DNJ8 A0A1B6HGY7 A0A1B6KIA3 A0A1Y1KIS3 A0A0N0BHC3 A0A1W4XSR9 U4UNZ9 N6U6C2 A0A1B6DHR8 A0A1B6CVS3 A0A1B6CPT4 E9HFH7 A0A0A9X2C0 A0A0P5LNR7 A0A0N7Z8H4 A0A2R7W4D1
A0A194PNC0 A0A067QWJ4 E2C5R7 A0A088AVX8 E9IUQ8 A0A310SVH7 F4WZP1 E2ACB4 A0A026X1C9 A0A3L8DRC8 K7IQY9 A0A2A3E2B1 A0A0L7QYB7 A0A195B7S1 A0A158NGT5 A0A195FFF8 A0A151XCW6 A0A195DXT7 A0A232F1V5 A0A1L8DSQ6 A0A154PR83 A0A1B0CDV0 D6W8B3 A0A1B6EQT1 A0A1Y1KNL2 A0A1B0DNJ8 A0A1B6HGY7 A0A1B6KIA3 A0A1Y1KIS3 A0A0N0BHC3 A0A1W4XSR9 U4UNZ9 N6U6C2 A0A1B6DHR8 A0A1B6CVS3 A0A1B6CPT4 E9HFH7 A0A0A9X2C0 A0A0P5LNR7 A0A0N7Z8H4 A0A2R7W4D1
Pubmed
EMBL
BABH01007788
EF554691
ABQ51915.1
KZ150007
PZC75189.1
NWSH01000604
+ More
PCG75368.1 ODYU01000355 SOQ34977.1 AGBW02010786 OWR47715.1 KQ461150 KPJ08743.1 KQ459597 KPI94931.1 KK852936 KDR13603.1 GL452801 EFN76701.1 GL766012 EFZ15691.1 KQ760118 OAD61923.1 GL888480 EGI60316.1 GL438491 EFN68904.1 KK107063 EZA61179.1 QOIP01000005 RLU22842.1 KZ288430 PBC25835.1 KQ414695 KOC63541.1 KQ976574 KYM80244.1 ADTU01015205 KQ981625 KYN39118.1 KQ982294 KYQ58205.1 KQ980107 KYN17688.1 NNAY01001282 OXU24500.1 GFDF01004680 JAV09404.1 KQ435073 KZC14402.1 AJWK01008274 KQ971307 EFA10926.2 GECZ01029511 JAS40258.1 GEZM01082840 JAV61255.1 AJVK01017614 GECU01033732 JAS73974.1 GEBQ01028796 JAT11181.1 GEZM01082841 JAV61254.1 KQ435757 KOX75848.1 KB632384 ERL94223.1 APGK01047053 KB741077 ENN74092.1 GEDC01012072 JAS25226.1 GEDC01019771 JAS17527.1 GEDC01021914 GEDC01017085 JAS15384.1 JAS20213.1 GL732636 EFX69504.1 GBHO01028727 GBRD01008581 GDHC01018525 JAG14877.1 JAG57240.1 JAQ00104.1 GDIQ01167281 JAK84444.1 GDKW01003339 JAI53256.1 KK854317 PTY14583.1
PCG75368.1 ODYU01000355 SOQ34977.1 AGBW02010786 OWR47715.1 KQ461150 KPJ08743.1 KQ459597 KPI94931.1 KK852936 KDR13603.1 GL452801 EFN76701.1 GL766012 EFZ15691.1 KQ760118 OAD61923.1 GL888480 EGI60316.1 GL438491 EFN68904.1 KK107063 EZA61179.1 QOIP01000005 RLU22842.1 KZ288430 PBC25835.1 KQ414695 KOC63541.1 KQ976574 KYM80244.1 ADTU01015205 KQ981625 KYN39118.1 KQ982294 KYQ58205.1 KQ980107 KYN17688.1 NNAY01001282 OXU24500.1 GFDF01004680 JAV09404.1 KQ435073 KZC14402.1 AJWK01008274 KQ971307 EFA10926.2 GECZ01029511 JAS40258.1 GEZM01082840 JAV61255.1 AJVK01017614 GECU01033732 JAS73974.1 GEBQ01028796 JAT11181.1 GEZM01082841 JAV61254.1 KQ435757 KOX75848.1 KB632384 ERL94223.1 APGK01047053 KB741077 ENN74092.1 GEDC01012072 JAS25226.1 GEDC01019771 JAS17527.1 GEDC01021914 GEDC01017085 JAS15384.1 JAS20213.1 GL732636 EFX69504.1 GBHO01028727 GBRD01008581 GDHC01018525 JAG14877.1 JAG57240.1 JAQ00104.1 GDIQ01167281 JAK84444.1 GDKW01003339 JAI53256.1 KK854317 PTY14583.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000027135
+ More
UP000008237 UP000005203 UP000007755 UP000000311 UP000053097 UP000279307 UP000002358 UP000242457 UP000053825 UP000078540 UP000005205 UP000078541 UP000075809 UP000078492 UP000215335 UP000076502 UP000092461 UP000007266 UP000092462 UP000053105 UP000192223 UP000030742 UP000019118 UP000000305
UP000008237 UP000005203 UP000007755 UP000000311 UP000053097 UP000279307 UP000002358 UP000242457 UP000053825 UP000078540 UP000005205 UP000078541 UP000075809 UP000078492 UP000215335 UP000076502 UP000092461 UP000007266 UP000092462 UP000053105 UP000192223 UP000030742 UP000019118 UP000000305
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A5JPL8
A0A2W1BJG9
A0A2A4JUJ6
A0A2H1V2A6
A0A212F1U9
A0A194QT92
+ More
A0A194PNC0 A0A067QWJ4 E2C5R7 A0A088AVX8 E9IUQ8 A0A310SVH7 F4WZP1 E2ACB4 A0A026X1C9 A0A3L8DRC8 K7IQY9 A0A2A3E2B1 A0A0L7QYB7 A0A195B7S1 A0A158NGT5 A0A195FFF8 A0A151XCW6 A0A195DXT7 A0A232F1V5 A0A1L8DSQ6 A0A154PR83 A0A1B0CDV0 D6W8B3 A0A1B6EQT1 A0A1Y1KNL2 A0A1B0DNJ8 A0A1B6HGY7 A0A1B6KIA3 A0A1Y1KIS3 A0A0N0BHC3 A0A1W4XSR9 U4UNZ9 N6U6C2 A0A1B6DHR8 A0A1B6CVS3 A0A1B6CPT4 E9HFH7 A0A0A9X2C0 A0A0P5LNR7 A0A0N7Z8H4 A0A2R7W4D1
A0A194PNC0 A0A067QWJ4 E2C5R7 A0A088AVX8 E9IUQ8 A0A310SVH7 F4WZP1 E2ACB4 A0A026X1C9 A0A3L8DRC8 K7IQY9 A0A2A3E2B1 A0A0L7QYB7 A0A195B7S1 A0A158NGT5 A0A195FFF8 A0A151XCW6 A0A195DXT7 A0A232F1V5 A0A1L8DSQ6 A0A154PR83 A0A1B0CDV0 D6W8B3 A0A1B6EQT1 A0A1Y1KNL2 A0A1B0DNJ8 A0A1B6HGY7 A0A1B6KIA3 A0A1Y1KIS3 A0A0N0BHC3 A0A1W4XSR9 U4UNZ9 N6U6C2 A0A1B6DHR8 A0A1B6CVS3 A0A1B6CPT4 E9HFH7 A0A0A9X2C0 A0A0P5LNR7 A0A0N7Z8H4 A0A2R7W4D1
PDB
4B86
E-value=1.00015e-28,
Score=316
Ontologies
GO
PANTHER
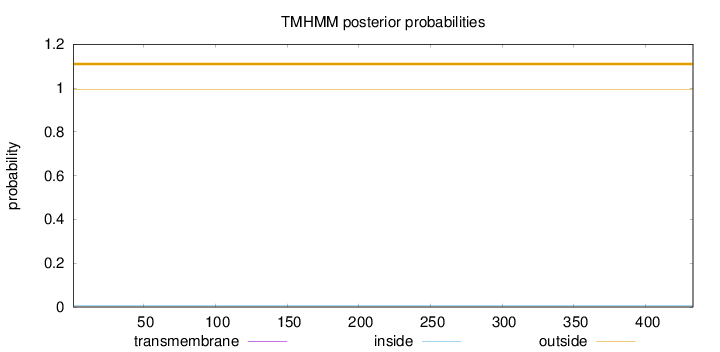
Topology
Length:
433
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00214
Exp number, first 60 AAs:
0.00114
Total prob of N-in:
0.00757
outside
1 - 433
Population Genetic Test Statistics
Pi
31.786815
Theta
23.552273
Tajima's D
1.219365
CLR
0.760252
CSRT
0.72256387180641
Interpretation
Uncertain
