Gene
KWMTBOMO02809
Annotation
reverse_transcriptase_[Anopheles_gambiae]
Location in the cell
Mitochondrial Reliability : 2.466
Sequence
CDS
ATGGTCACATTCTGGCGCAACTTGTGGTCAAGACCTGTGGAGCATGTGGAGGGCTCCTGGATGCAGGTCATTGAGGACTCGTGTGCGGGCATACCACCGATGAGCCCGGTTACTATCAGCAAAGGTGATATTAAGGCTGCAGTCTGTAGTTCTTCACACTGGAAGTCTCCAGGCTGCGACGGACTGCAGCTTTACTGGCTAAAAAGTTTCCGGGCTTGTCACGAAACCCTCGCCAGACAGTTCCAGGAAGCCCTAGATACAAAAGTGCTCCCCGCCTTCCTCACTACTGGGATCACTCACCTCATTCCAAAATCGGAAAGTACCGCGGATCCGGCACAGTACCGGCCAATAACGTGTCTTCCTACCACCTATAAGACACTGACATCCGTTCTGGAAACTAAGATCTCACACCACATAGACAGCTGTCGAGTACTGTCTGGTGCCCAGAACGGGTGTAGGCGTGGTAGCCGCGGTACTAAGGAACTCCTCTTGATCGACGCGGTAGCTGGCCAGCAGGTCAAACGCAACCGACGAAATTTCTCTGCCGCCTGGATAGATTATAAAAAGGCATTTGACTCGGTCCCCCATACATGGCTGAAGAGGGTCCTCGAGCTGTATAAGATAGACGATACAGTTCGAGACTTCCTCGGTGCCTGTATGGGGCAATGGAGTACAATGCTTAGTCTATCCGGTGTGCGGCTGTCGGCTGTAGAAGACCGAATAAAGGTGAATCGGGGTATATTTCAGGGTGATTGTCTGAGTCCATTGTGGTTTTGCCTGGCCCTGAATCCTCTCAGTTCATTACTGGAGGCTTCGAGGACAGGCTTTCAGTTCCGAAGGGGGGGCTTGAAGGTGTCCCACTTGTTCTACATGGATGACCTCAAGTTGTACGCTCCTCATGCTGCAGGCCTCAGGCGACTGCTTGACATCACTTGTGATTTCAGTAGTCGCATTAGGATGAGGCTGGGAGTCGACAAGTGTGCGGTTCTCCATGTAGAGAGAGGGAGCATAGTCTCATCGGATCGTTTACCTGTATCCGATGGGATAAACCTTCGGGCACTCTCCGAGGGAGAAACATACCGGTACCTGGGCATGTCGCAAAATTTAGGTATTGACGTGGCGGTCGTCAAACAAGCTGCATGCGACGTGTTTTATGAACGTTTGACAAAAGTGTGTAAGAGCCTTTTGTCGGGCGTGAATAAGACAAGGGCGTTTAACGGCTGGGCGATGCCCGTCCTGTTGTATACCTTCGGCGTGCTCAGGTGGACTCAGACCGAACTGGATGCCCTGGATAGAAAAGTCCGCTCAATTATGACCCTCCATAGGATGCTCCACCCGAAGTCCTCGGTAATGAGATTGTACATTCCGCGGAAGTGTGGTGGCCGGGGCCTACTTAGTGCCAAGGTCATGCACAACAGTGGGGTGTGCAGTCTCAGGGAATATTTCCTATCGAGGGCTGACAGTGTGATGCACAGAGAGGTCATGGAATGCGACAAAGGACTGACCCCGCTAGCCCTGGCCAGAGACGGGTGGCAATCACCGGCGGTACTCAGTACCTCTGACCGTGAGGCCATATGGAAAGGGAAAGAGCTGCACGGACGCTTCTTTCAGGCACTGCATGAGCCCCACGTGGACAAAGAAGCCTCCGTGCACTGGCTGCGATTCGGTGACCTCTTTGGGGAAACCGAGGGTTTTGTCTGTGCAATACAGGATCAGGTTATCAGGACGAACAACTATAGGAAGCACATTCTGAGGGATGGGACAGCTGACATCTGTCGATTATGCCGCCGACCGGGTGAATCCCTCAGACATGTCACCTCTGGTTGTTCTATGCTTGCTAACACTGAGTACTTGCACAGACATAACCAAGCAGCCAGAATCCTCCACCAAGAGCTCGCCCTTAAGTATGGCCTCATCGAACAGAGGCTACCGTATTATAAGTACCAGCCGGACGCTGTACTCGAAAACGACCGTGCCAAGCTCTACTGGGACCGGCCCATCATTACGGACAGGACTATTCTTGCGAATAAGCCTGATATCGTGCTGATGGACCGGACGGAGTCTCGGGTATTTCTGGTGGATATCACCATCCCCTACGACGAGAACCTCGTGCGGGCCGAGGCAGATAAAAAGACCAAATATTTGGACCTGGCGCACGAGGTGACCGACATGTGGAGGGTGGTATCTACAGAAATAATCCCGGTAGTTGTGTCGGTGAATGGTTTGGTCCCTAAAAGCCTCTCAAAACATCTCGAGAGGCTTGGTCTCAACAAAAAGTCGGTGGTGGCCCAAATGCAAAAGGCAGTCTTGCTCGATAATGCCCGTATAGTTCGCCGGTTTCTCTCCCAATAG
Protein
MVTFWRNLWSRPVEHVEGSWMQVIEDSCAGIPPMSPVTISKGDIKAAVCSSSHWKSPGCDGLQLYWLKSFRACHETLARQFQEALDTKVLPAFLTTGITHLIPKSESTADPAQYRPITCLPTTYKTLTSVLETKISHHIDSCRVLSGAQNGCRRGSRGTKELLLIDAVAGQQVKRNRRNFSAAWIDYKKAFDSVPHTWLKRVLELYKIDDTVRDFLGACMGQWSTMLSLSGVRLSAVEDRIKVNRGIFQGDCLSPLWFCLALNPLSSLLEASRTGFQFRRGGLKVSHLFYMDDLKLYAPHAAGLRRLLDITCDFSSRIRMRLGVDKCAVLHVERGSIVSSDRLPVSDGINLRALSEGETYRYLGMSQNLGIDVAVVKQAACDVFYERLTKVCKSLLSGVNKTRAFNGWAMPVLLYTFGVLRWTQTELDALDRKVRSIMTLHRMLHPKSSVMRLYIPRKCGGRGLLSAKVMHNSGVCSLREYFLSRADSVMHREVMECDKGLTPLALARDGWQSPAVLSTSDREAIWKGKELHGRFFQALHEPHVDKEASVHWLRFGDLFGETEGFVCAIQDQVIRTNNYRKHILRDGTADICRLCRRPGESLRHVTSGCSMLANTEYLHRHNQAARILHQELALKYGLIEQRLPYYKYQPDAVLENDRAKLYWDRPIITDRTILANKPDIVLMDRTESRVFLVDITIPYDENLVRAEADKKTKYLDLAHEVTDMWRVVSTEIIPVVVSVNGLVPKSLSKHLERLGLNKKSVVAQMQKAVLLDNARIVRRFLSQ
Summary
Uniprot
A0A1S4EQ35
W8BF34
Q76IN5
W8AXW6
D7GYL6
A0A2J7PDQ6
+ More
A0A2J7PIT9 E0VPJ4 E0VSD9 E0W0M4 E0W0H3 E0VPK6 E0VA19 A0A3B3I0I7 H3AB60 A0A3P9MNJ5 A0A3B3HW39 A0A3P9MMW4 A0A3P9LVD0 A0A3P8NVY4 A0A3P8NT65 A0A3B3H9G8 A0A3P9K6Z6 A0A3B3HU40 A0A3P8NFL6 A0A3B3ILW9 A0A3P8P1Q8 A0A3P8QC13 A0A3P8PQZ9 A0A3P8PU69 A0A3B3HQX9 A0A3P8R5E8 A0A3P9M4C8 A0A3P8QAR5 A0A3P8Q3U1 A0A3B3I5C9 A0A3P8NKM6 A0A3P8PU63 A0A3P8PST2 A0A3B3H8G1 A0A3P8QTM2 A0A3P8QPZ9 A0A3P8PSV9 A0A3B3HNT5 A0A3P8Q816 A0A3P8NRB8 A0A3P8QK92 A0A3P8QRC6 A0A3P8P1I6 A0A3P8PUT7 A0A3P9DUW1 A0A3P8P8J8 A0A3P8P9Q9 A0A3P8Q8N4 A0A3P8PIV2 A0A3P8NH43 A0A3P8NLS5 A0A3P8QJR1 A0A3B3HQ99 A0A3P9QFG6 A0A3B5PPD0 H3BBL0 A0A3P8QRQ3 A0A0S7ESG3 A0A3B3H9C1 A0A2P2HZD8 H3AYE9 A0A3P8RI52 H3B0S5 A0A3P8Q6B2 A0A3P9LCQ1 A0A1X7SMI5 H3A710 A0A3Q0J2S4 H3A541 H3AWP0
A0A2J7PIT9 E0VPJ4 E0VSD9 E0W0M4 E0W0H3 E0VPK6 E0VA19 A0A3B3I0I7 H3AB60 A0A3P9MNJ5 A0A3B3HW39 A0A3P9MMW4 A0A3P9LVD0 A0A3P8NVY4 A0A3P8NT65 A0A3B3H9G8 A0A3P9K6Z6 A0A3B3HU40 A0A3P8NFL6 A0A3B3ILW9 A0A3P8P1Q8 A0A3P8QC13 A0A3P8PQZ9 A0A3P8PU69 A0A3B3HQX9 A0A3P8R5E8 A0A3P9M4C8 A0A3P8QAR5 A0A3P8Q3U1 A0A3B3I5C9 A0A3P8NKM6 A0A3P8PU63 A0A3P8PST2 A0A3B3H8G1 A0A3P8QTM2 A0A3P8QPZ9 A0A3P8PSV9 A0A3B3HNT5 A0A3P8Q816 A0A3P8NRB8 A0A3P8QK92 A0A3P8QRC6 A0A3P8P1I6 A0A3P8PUT7 A0A3P9DUW1 A0A3P8P8J8 A0A3P8P9Q9 A0A3P8Q8N4 A0A3P8PIV2 A0A3P8NH43 A0A3P8NLS5 A0A3P8QJR1 A0A3B3HQ99 A0A3P9QFG6 A0A3B5PPD0 H3BBL0 A0A3P8QRQ3 A0A0S7ESG3 A0A3B3H9C1 A0A2P2HZD8 H3AYE9 A0A3P8RI52 H3B0S5 A0A3P8Q6B2 A0A3P9LCQ1 A0A1X7SMI5 H3A710 A0A3Q0J2S4 H3A541 H3AWP0
EMBL
GAMC01018221
JAB88334.1
AB097127
BAC82595.1
GAMC01016897
JAB89658.1
+ More
GG695857 EFA13462.1 NEVH01026385 PNF14463.1 NEVH01024955 PNF16260.1 DS235366 EEB15300.1 DS235750 EEB16295.1 DS235861 EEB19180.1 DS235859 EEB19129.1 EEB15312.1 DS235004 EEB10225.1 AFYH01227415 AFYH01052121 GBYX01473740 JAO07919.1 IACF01001441 LAB67141.1 AFYH01132769 AFYH01092381 AFYH01092382 AFYH01245796 AFYH01248595 AFYH01142702 AFYH01142703
GG695857 EFA13462.1 NEVH01026385 PNF14463.1 NEVH01024955 PNF16260.1 DS235366 EEB15300.1 DS235750 EEB16295.1 DS235861 EEB19180.1 DS235859 EEB19129.1 EEB15312.1 DS235004 EEB10225.1 AFYH01227415 AFYH01052121 GBYX01473740 JAO07919.1 IACF01001441 LAB67141.1 AFYH01132769 AFYH01092381 AFYH01092382 AFYH01245796 AFYH01248595 AFYH01142702 AFYH01142703
Proteomes
Pfam
PF00078 RVT_1
ProteinModelPortal
A0A1S4EQ35
W8BF34
Q76IN5
W8AXW6
D7GYL6
A0A2J7PDQ6
+ More
A0A2J7PIT9 E0VPJ4 E0VSD9 E0W0M4 E0W0H3 E0VPK6 E0VA19 A0A3B3I0I7 H3AB60 A0A3P9MNJ5 A0A3B3HW39 A0A3P9MMW4 A0A3P9LVD0 A0A3P8NVY4 A0A3P8NT65 A0A3B3H9G8 A0A3P9K6Z6 A0A3B3HU40 A0A3P8NFL6 A0A3B3ILW9 A0A3P8P1Q8 A0A3P8QC13 A0A3P8PQZ9 A0A3P8PU69 A0A3B3HQX9 A0A3P8R5E8 A0A3P9M4C8 A0A3P8QAR5 A0A3P8Q3U1 A0A3B3I5C9 A0A3P8NKM6 A0A3P8PU63 A0A3P8PST2 A0A3B3H8G1 A0A3P8QTM2 A0A3P8QPZ9 A0A3P8PSV9 A0A3B3HNT5 A0A3P8Q816 A0A3P8NRB8 A0A3P8QK92 A0A3P8QRC6 A0A3P8P1I6 A0A3P8PUT7 A0A3P9DUW1 A0A3P8P8J8 A0A3P8P9Q9 A0A3P8Q8N4 A0A3P8PIV2 A0A3P8NH43 A0A3P8NLS5 A0A3P8QJR1 A0A3B3HQ99 A0A3P9QFG6 A0A3B5PPD0 H3BBL0 A0A3P8QRQ3 A0A0S7ESG3 A0A3B3H9C1 A0A2P2HZD8 H3AYE9 A0A3P8RI52 H3B0S5 A0A3P8Q6B2 A0A3P9LCQ1 A0A1X7SMI5 H3A710 A0A3Q0J2S4 H3A541 H3AWP0
A0A2J7PIT9 E0VPJ4 E0VSD9 E0W0M4 E0W0H3 E0VPK6 E0VA19 A0A3B3I0I7 H3AB60 A0A3P9MNJ5 A0A3B3HW39 A0A3P9MMW4 A0A3P9LVD0 A0A3P8NVY4 A0A3P8NT65 A0A3B3H9G8 A0A3P9K6Z6 A0A3B3HU40 A0A3P8NFL6 A0A3B3ILW9 A0A3P8P1Q8 A0A3P8QC13 A0A3P8PQZ9 A0A3P8PU69 A0A3B3HQX9 A0A3P8R5E8 A0A3P9M4C8 A0A3P8QAR5 A0A3P8Q3U1 A0A3B3I5C9 A0A3P8NKM6 A0A3P8PU63 A0A3P8PST2 A0A3B3H8G1 A0A3P8QTM2 A0A3P8QPZ9 A0A3P8PSV9 A0A3B3HNT5 A0A3P8Q816 A0A3P8NRB8 A0A3P8QK92 A0A3P8QRC6 A0A3P8P1I6 A0A3P8PUT7 A0A3P9DUW1 A0A3P8P8J8 A0A3P8P9Q9 A0A3P8Q8N4 A0A3P8PIV2 A0A3P8NH43 A0A3P8NLS5 A0A3P8QJR1 A0A3B3HQ99 A0A3P9QFG6 A0A3B5PPD0 H3BBL0 A0A3P8QRQ3 A0A0S7ESG3 A0A3B3H9C1 A0A2P2HZD8 H3AYE9 A0A3P8RI52 H3B0S5 A0A3P8Q6B2 A0A3P9LCQ1 A0A1X7SMI5 H3A710 A0A3Q0J2S4 H3A541 H3AWP0
Ontologies
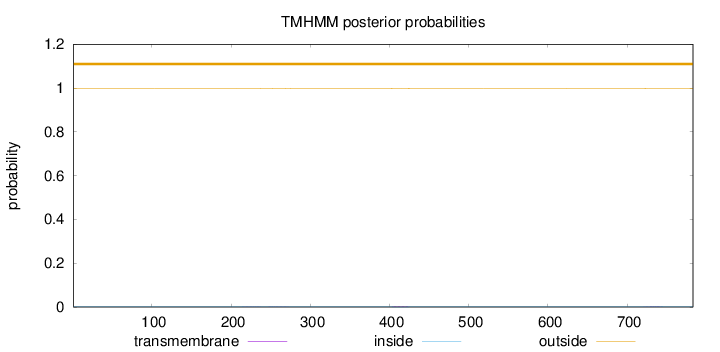
Topology
Length:
783
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0910499999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00198
outside
1 - 783
Population Genetic Test Statistics
Pi
180.837654
Theta
72.374744
Tajima's D
4.143026
CLR
0.453829
CSRT
0.99930003499825
Interpretation
Uncertain
