Gene
KWMTBOMO02805 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003507
Annotation
PREDICTED:_poly(U)-specific_endoribonuclease-D_isoform_X1_[Bombyx_mori]
Full name
Poly(U)-specific endoribonuclease homolog
Alternative Name
Protein endoU
Uridylate-specific endoribonuclease homolog
Uridylate-specific endoribonuclease homolog
Location in the cell
Mitochondrial Reliability : 1.378 Nuclear Reliability : 1.06
Sequence
CDS
ATGACACTGAGTGAAGAACTTCTTCGGAAAGATACAAACAATGCTGCCAAATATATCACGATGGACTTTCAAGAAAAAACGACATCACAGTCGAAAGAAGACAAAGCCCCGTTGCCGCTGCTAAATATATCATCAGCGGCATGGAATATTCCAACAATAGAAAAAATTGTCCCACTCTTGGACAATTACGAAAGAAGCACGTTAGTAAATGAACATGTAACGCCACAGGAACGAAACGAGGAAAATGCGTTTATGGACGCCATCATGGCCACAAGTGTTATGAGGCATTTAATGAATTTTTTGAAAGAAAAAGGTTATGTCACACCGGATCCTAGACAACAAAGAGATTTTGTAAAGCAGCTCTGGTTTAGTTTGTACTCGAGGGGAAAAGGAAAAATCAGTAGTTCAGGATTTGAACATATATTTGTATCTGAATTAAAAAATGGCGAAGTTTCTGGTCTACATAATTGGATTTATTTTTCAAAGGAAGAAGCAGCGAACCGACTGAATTACTTAGGTTACTTGAAATACAGACAATTTGGGGAGCGTGGAGCAGTGATTAAATTCCACTTCACCCAACAAGGAGTGGACAAGCCGGTTAACTCTATGTTCATTGGTACATCACCTGAGTTGGAAATGGCATTGTATACTTTGTGCTTCGTGACTCGTGCTGACAGCGAGTGTAACCTGAAACTGTCCAACAACGATGTCAGTATCATGACTCACACTTTCCGTTACAGGAGTAAAAATCTAATCGGTAGTGGCTATCCACAGATCTAA
Protein
MTLSEELLRKDTNNAAKYITMDFQEKTTSQSKEDKAPLPLLNISSAAWNIPTIEKIVPLLDNYERSTLVNEHVTPQERNEENAFMDAIMATSVMRHLMNFLKEKGYVTPDPRQQRDFVKQLWFSLYSRGKGKISSSGFEHIFVSELKNGEVSGLHNWIYFSKEEAANRLNYLGYLKYRQFGERGAVIKFHFTQQGVDKPVNSMFIGTSPELEMALYTLCFVTRADSECNLKLSNNDVSIMTHTFRYRSKNLIGSGYPQI
Summary
Description
Endoribonuclease.
Cofactor
Mn(2+)
Subunit
Monomer.
Similarity
Belongs to the MCM family.
Belongs to the ENDOU family.
Belongs to the ENDOU family.
Keywords
Complete proteome
Endonuclease
Hydrolase
Manganese
Metal-binding
Nuclease
Reference proteome
RNA-binding
Secreted
Signal
Feature
chain Poly(U)-specific endoribonuclease homolog
Uniprot
H9J1X0
A0A1E1WJY3
A0A2W1BPF9
S4P3D5
A0A194PPM0
I4DRI3
+ More
A0A194QUU4 I4DPG3 A0A2A4JV71 A0A2A4JVG1 A0A2P8Z2R2 U4UIW8 B4H4U2 N6TQT1 A0A067QX30 Q29I02 B3MQP8 A0A0M3QZI8 A0A3B0K5W6 D3TQ61 A0A1W4V1D5 B4M819 B4MSH8 A0A1A9ZEJ9 B4L628 A0A1B0FRI4 A0A0T6B131 A0A1A9XBT6 A0A1B0BDZ2 A0A0L0C871 D6WSA3 A0A1A9WT30 A0A336L5G4 B3NVC5 A0A1I8PMP8 A0A1I8PMU6 B4JKK2 A0A1A9VCK9 B4R2P9 A0A1I8M0F7 X2JJD6 Q9VZ49 A0A1J1HRH3 T1PE22 U5EP18 A0A034VRM2 W8B102 B4IDV9 A0A0J7MSF7 A0A232FJB6 K7IUV7 A0A0K8U9P9 A0A0K8W848 A0A1Y1LQD3 A0A034VP83 A0A0A1WPF2 A0A2M4BLL3 T1E1H9 A0A2M3YZ96 A0A2M4BJW7 A0A2M3YZB4 A0A1S4FMU8 A0A1W6EVV9 A0A3L8D6C2 A0A026VYP5 A0A182QLB6 A0A2M4AC62 A0A2M4ABY0 A0A182VRQ6 T1E7J1 A0A182JDA4 A0A2M4DP21 A0A182XV19 A0A0C9QL97 Q16VA7 A0A1L8DY31 A0A182F7B5 A0A1B6FV66 F5HKF0 B8RJ46 A0A182XHG2 A0A182TY62 A0A182GGQ1 A0A182LG51 A0A182HTY4 A0A023EV40 A0A182V8M2 A0A1B6HJW4 A0A023EU03 A0A023EV44 E2BWC0 B4PYV9 A0A1Q3FM77 A0A182N4S7 E9IR65 E2APY1 B0X4J0 A0A0L7RIF8 A0A182RVA7 E2BWB9
A0A194QUU4 I4DPG3 A0A2A4JV71 A0A2A4JVG1 A0A2P8Z2R2 U4UIW8 B4H4U2 N6TQT1 A0A067QX30 Q29I02 B3MQP8 A0A0M3QZI8 A0A3B0K5W6 D3TQ61 A0A1W4V1D5 B4M819 B4MSH8 A0A1A9ZEJ9 B4L628 A0A1B0FRI4 A0A0T6B131 A0A1A9XBT6 A0A1B0BDZ2 A0A0L0C871 D6WSA3 A0A1A9WT30 A0A336L5G4 B3NVC5 A0A1I8PMP8 A0A1I8PMU6 B4JKK2 A0A1A9VCK9 B4R2P9 A0A1I8M0F7 X2JJD6 Q9VZ49 A0A1J1HRH3 T1PE22 U5EP18 A0A034VRM2 W8B102 B4IDV9 A0A0J7MSF7 A0A232FJB6 K7IUV7 A0A0K8U9P9 A0A0K8W848 A0A1Y1LQD3 A0A034VP83 A0A0A1WPF2 A0A2M4BLL3 T1E1H9 A0A2M3YZ96 A0A2M4BJW7 A0A2M3YZB4 A0A1S4FMU8 A0A1W6EVV9 A0A3L8D6C2 A0A026VYP5 A0A182QLB6 A0A2M4AC62 A0A2M4ABY0 A0A182VRQ6 T1E7J1 A0A182JDA4 A0A2M4DP21 A0A182XV19 A0A0C9QL97 Q16VA7 A0A1L8DY31 A0A182F7B5 A0A1B6FV66 F5HKF0 B8RJ46 A0A182XHG2 A0A182TY62 A0A182GGQ1 A0A182LG51 A0A182HTY4 A0A023EV40 A0A182V8M2 A0A1B6HJW4 A0A023EU03 A0A023EV44 E2BWC0 B4PYV9 A0A1Q3FM77 A0A182N4S7 E9IR65 E2APY1 B0X4J0 A0A0L7RIF8 A0A182RVA7 E2BWB9
EC Number
3.1.-.-
Pubmed
19121390
28756777
23622113
26354079
22651552
29403074
+ More
23537049 17994087 24845553 15632085 20353571 26108605 18362917 19820115 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 25348373 24495485 28648823 20075255 28004739 25830018 24330624 30249741 24508170 25244985 17510324 12364791 14747013 17210077 26483478 20966253 24945155 20798317 17550304 21282665
23537049 17994087 24845553 15632085 20353571 26108605 18362917 19820115 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 25348373 24495485 28648823 20075255 28004739 25830018 24330624 30249741 24508170 25244985 17510324 12364791 14747013 17210077 26483478 20966253 24945155 20798317 17550304 21282665
EMBL
BABH01007795
BABH01007796
BABH01007797
BABH01007798
GDQN01003725
JAT87329.1
+ More
KZ150007 PZC75197.1 GAIX01006489 JAA86071.1 KQ459597 KPI94923.1 AK405130 BAM20523.1 KQ461150 KPJ08750.1 AK403585 BAM19803.1 NWSH01000604 PCG75383.1 PCG75382.1 PYGN01000223 PSN50777.1 KB632322 ERL92343.1 CH479209 EDW32704.1 APGK01058327 KB741288 ENN70666.1 KK853256 KDR09265.1 CH379064 EAL31604.3 CH902621 EDV44674.2 CP012528 ALC49493.1 OUUW01000025 SPP89617.1 EZ423563 ADD19839.1 CH940653 EDW62295.1 CH963851 EDW75067.1 CH933812 EDW05824.1 CCAG010017603 LJIG01016277 KRT81084.1 JXJN01012729 JRES01000767 KNC28456.1 KQ971354 EFA05915.2 UFQS01001083 UFQT01001083 SSX08890.1 SSX28801.1 CH954180 EDV46113.1 CH916370 EDW00105.1 CM000366 EDX17598.1 AE014298 AHN59572.1 BT031314 AY075327 CVRI01000016 CRK90130.1 KA646954 AFP61583.1 GANO01003858 JAB56013.1 GAKP01013838 JAC45114.1 GAMC01011800 JAB94755.1 CH480830 EDW45767.1 LBMM01019810 KMQ83445.1 NNAY01000115 OXU30836.1 AAZX01008132 GDHF01029234 JAI23080.1 GDHF01005259 JAI47055.1 GEZM01049779 GEZM01049778 JAV75853.1 GAKP01013836 JAC45116.1 GBXI01013565 JAD00727.1 GGFJ01004781 MBW53922.1 GALA01001649 JAA93203.1 GGFM01000848 MBW21599.1 GGFJ01004225 MBW53366.1 GGFM01000830 MBW21581.1 KY563451 ARK19860.1 QOIP01000012 RLU15786.1 KK107565 EZA48923.1 AXCN02000015 GGFK01004991 MBW38312.1 GGFK01004948 MBW38269.1 GAMD01002997 JAA98593.1 GGFL01015139 MBW79317.1 GBYB01004299 JAG74066.1 CH477598 EAT38484.1 GFDF01002782 JAV11302.1 GECZ01015715 JAS54054.1 AAAB01008859 EAA07623.5 EGK96701.1 EGK96702.1 EZ000471 ACJ64345.1 JXUM01062154 JXUM01062155 JXUM01062156 KQ562186 KXJ76466.1 APCN01000591 GAPW01000737 JAC12861.1 GECU01032712 GECU01015858 JAS74994.1 JAS91848.1 GAPW01000738 JAC12860.1 GAPW01000721 JAC12877.1 GL451103 EFN80024.1 CM000162 EDX02037.1 GFDL01006364 JAV28681.1 GL765088 EFZ16938.1 GL441630 EFN64513.1 DS232343 EDS40407.1 KQ414584 KOC70614.1 EFN80023.1
KZ150007 PZC75197.1 GAIX01006489 JAA86071.1 KQ459597 KPI94923.1 AK405130 BAM20523.1 KQ461150 KPJ08750.1 AK403585 BAM19803.1 NWSH01000604 PCG75383.1 PCG75382.1 PYGN01000223 PSN50777.1 KB632322 ERL92343.1 CH479209 EDW32704.1 APGK01058327 KB741288 ENN70666.1 KK853256 KDR09265.1 CH379064 EAL31604.3 CH902621 EDV44674.2 CP012528 ALC49493.1 OUUW01000025 SPP89617.1 EZ423563 ADD19839.1 CH940653 EDW62295.1 CH963851 EDW75067.1 CH933812 EDW05824.1 CCAG010017603 LJIG01016277 KRT81084.1 JXJN01012729 JRES01000767 KNC28456.1 KQ971354 EFA05915.2 UFQS01001083 UFQT01001083 SSX08890.1 SSX28801.1 CH954180 EDV46113.1 CH916370 EDW00105.1 CM000366 EDX17598.1 AE014298 AHN59572.1 BT031314 AY075327 CVRI01000016 CRK90130.1 KA646954 AFP61583.1 GANO01003858 JAB56013.1 GAKP01013838 JAC45114.1 GAMC01011800 JAB94755.1 CH480830 EDW45767.1 LBMM01019810 KMQ83445.1 NNAY01000115 OXU30836.1 AAZX01008132 GDHF01029234 JAI23080.1 GDHF01005259 JAI47055.1 GEZM01049779 GEZM01049778 JAV75853.1 GAKP01013836 JAC45116.1 GBXI01013565 JAD00727.1 GGFJ01004781 MBW53922.1 GALA01001649 JAA93203.1 GGFM01000848 MBW21599.1 GGFJ01004225 MBW53366.1 GGFM01000830 MBW21581.1 KY563451 ARK19860.1 QOIP01000012 RLU15786.1 KK107565 EZA48923.1 AXCN02000015 GGFK01004991 MBW38312.1 GGFK01004948 MBW38269.1 GAMD01002997 JAA98593.1 GGFL01015139 MBW79317.1 GBYB01004299 JAG74066.1 CH477598 EAT38484.1 GFDF01002782 JAV11302.1 GECZ01015715 JAS54054.1 AAAB01008859 EAA07623.5 EGK96701.1 EGK96702.1 EZ000471 ACJ64345.1 JXUM01062154 JXUM01062155 JXUM01062156 KQ562186 KXJ76466.1 APCN01000591 GAPW01000737 JAC12861.1 GECU01032712 GECU01015858 JAS74994.1 JAS91848.1 GAPW01000738 JAC12860.1 GAPW01000721 JAC12877.1 GL451103 EFN80024.1 CM000162 EDX02037.1 GFDL01006364 JAV28681.1 GL765088 EFZ16938.1 GL441630 EFN64513.1 DS232343 EDS40407.1 KQ414584 KOC70614.1 EFN80023.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000245037
UP000030742
+ More
UP000008744 UP000019118 UP000027135 UP000001819 UP000007801 UP000092553 UP000268350 UP000192221 UP000008792 UP000007798 UP000092445 UP000009192 UP000092444 UP000092443 UP000092460 UP000037069 UP000007266 UP000091820 UP000008711 UP000095300 UP000001070 UP000078200 UP000000304 UP000095301 UP000000803 UP000183832 UP000001292 UP000036403 UP000215335 UP000002358 UP000279307 UP000053097 UP000075886 UP000075920 UP000075880 UP000076408 UP000008820 UP000069272 UP000007062 UP000076407 UP000075902 UP000069940 UP000249989 UP000075882 UP000075840 UP000075903 UP000008237 UP000002282 UP000075884 UP000000311 UP000002320 UP000053825 UP000075900
UP000008744 UP000019118 UP000027135 UP000001819 UP000007801 UP000092553 UP000268350 UP000192221 UP000008792 UP000007798 UP000092445 UP000009192 UP000092444 UP000092443 UP000092460 UP000037069 UP000007266 UP000091820 UP000008711 UP000095300 UP000001070 UP000078200 UP000000304 UP000095301 UP000000803 UP000183832 UP000001292 UP000036403 UP000215335 UP000002358 UP000279307 UP000053097 UP000075886 UP000075920 UP000075880 UP000076408 UP000008820 UP000069272 UP000007062 UP000076407 UP000075902 UP000069940 UP000249989 UP000075882 UP000075840 UP000075903 UP000008237 UP000002282 UP000075884 UP000000311 UP000002320 UP000053825 UP000075900
Interpro
Gene 3D
ProteinModelPortal
H9J1X0
A0A1E1WJY3
A0A2W1BPF9
S4P3D5
A0A194PPM0
I4DRI3
+ More
A0A194QUU4 I4DPG3 A0A2A4JV71 A0A2A4JVG1 A0A2P8Z2R2 U4UIW8 B4H4U2 N6TQT1 A0A067QX30 Q29I02 B3MQP8 A0A0M3QZI8 A0A3B0K5W6 D3TQ61 A0A1W4V1D5 B4M819 B4MSH8 A0A1A9ZEJ9 B4L628 A0A1B0FRI4 A0A0T6B131 A0A1A9XBT6 A0A1B0BDZ2 A0A0L0C871 D6WSA3 A0A1A9WT30 A0A336L5G4 B3NVC5 A0A1I8PMP8 A0A1I8PMU6 B4JKK2 A0A1A9VCK9 B4R2P9 A0A1I8M0F7 X2JJD6 Q9VZ49 A0A1J1HRH3 T1PE22 U5EP18 A0A034VRM2 W8B102 B4IDV9 A0A0J7MSF7 A0A232FJB6 K7IUV7 A0A0K8U9P9 A0A0K8W848 A0A1Y1LQD3 A0A034VP83 A0A0A1WPF2 A0A2M4BLL3 T1E1H9 A0A2M3YZ96 A0A2M4BJW7 A0A2M3YZB4 A0A1S4FMU8 A0A1W6EVV9 A0A3L8D6C2 A0A026VYP5 A0A182QLB6 A0A2M4AC62 A0A2M4ABY0 A0A182VRQ6 T1E7J1 A0A182JDA4 A0A2M4DP21 A0A182XV19 A0A0C9QL97 Q16VA7 A0A1L8DY31 A0A182F7B5 A0A1B6FV66 F5HKF0 B8RJ46 A0A182XHG2 A0A182TY62 A0A182GGQ1 A0A182LG51 A0A182HTY4 A0A023EV40 A0A182V8M2 A0A1B6HJW4 A0A023EU03 A0A023EV44 E2BWC0 B4PYV9 A0A1Q3FM77 A0A182N4S7 E9IR65 E2APY1 B0X4J0 A0A0L7RIF8 A0A182RVA7 E2BWB9
A0A194QUU4 I4DPG3 A0A2A4JV71 A0A2A4JVG1 A0A2P8Z2R2 U4UIW8 B4H4U2 N6TQT1 A0A067QX30 Q29I02 B3MQP8 A0A0M3QZI8 A0A3B0K5W6 D3TQ61 A0A1W4V1D5 B4M819 B4MSH8 A0A1A9ZEJ9 B4L628 A0A1B0FRI4 A0A0T6B131 A0A1A9XBT6 A0A1B0BDZ2 A0A0L0C871 D6WSA3 A0A1A9WT30 A0A336L5G4 B3NVC5 A0A1I8PMP8 A0A1I8PMU6 B4JKK2 A0A1A9VCK9 B4R2P9 A0A1I8M0F7 X2JJD6 Q9VZ49 A0A1J1HRH3 T1PE22 U5EP18 A0A034VRM2 W8B102 B4IDV9 A0A0J7MSF7 A0A232FJB6 K7IUV7 A0A0K8U9P9 A0A0K8W848 A0A1Y1LQD3 A0A034VP83 A0A0A1WPF2 A0A2M4BLL3 T1E1H9 A0A2M3YZ96 A0A2M4BJW7 A0A2M3YZB4 A0A1S4FMU8 A0A1W6EVV9 A0A3L8D6C2 A0A026VYP5 A0A182QLB6 A0A2M4AC62 A0A2M4ABY0 A0A182VRQ6 T1E7J1 A0A182JDA4 A0A2M4DP21 A0A182XV19 A0A0C9QL97 Q16VA7 A0A1L8DY31 A0A182F7B5 A0A1B6FV66 F5HKF0 B8RJ46 A0A182XHG2 A0A182TY62 A0A182GGQ1 A0A182LG51 A0A182HTY4 A0A023EV40 A0A182V8M2 A0A1B6HJW4 A0A023EU03 A0A023EV44 E2BWC0 B4PYV9 A0A1Q3FM77 A0A182N4S7 E9IR65 E2APY1 B0X4J0 A0A0L7RIF8 A0A182RVA7 E2BWB9
PDB
2C1W
E-value=4.41454e-30,
Score=325
Ontologies
GO
Topology
Subcellular location
Secreted
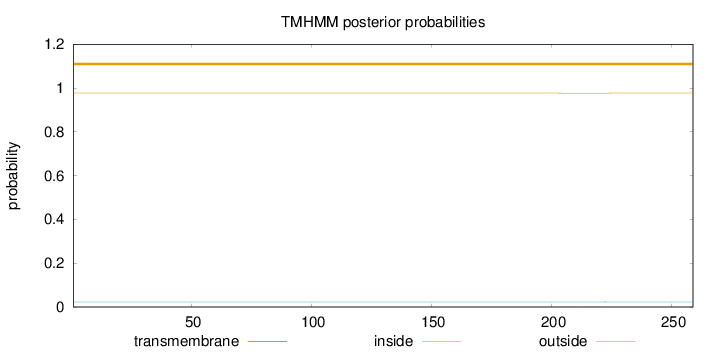
Length:
259
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00556
Exp number, first 60 AAs:
0.00079
Total prob of N-in:
0.02307
outside
1 - 259
Population Genetic Test Statistics
Pi
239.448203
Theta
185.707481
Tajima's D
0.63077
CLR
0.642383
CSRT
0.560221988900555
Interpretation
Uncertain
