Gene
KWMTBOMO02804
Annotation
PREDICTED:_protein-cysteine_N-palmitoyltransferase_HHAT-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.312
Sequence
CDS
ATGTCTGAAACACTGGCTATAAAAAGTTTGCCGACATTAGCTCTTTGTGGAGGTGGTTTATGGATGGGTTTAGTATTCCATTTAAAGTATGTCATATCATATGGCACAACAGGAGCTTTCGCCAGATTAGACAATATGGAACCACCACCAATTCCAAGATGTGTTGCAAGAATTCATGTCTATTCTCTCATGTGGAGATATTTTGATGTTGGATTATACCAGTTTTTATCCAAATATATCTACAAACCTGGTTACAGTGTCCTATCTAATTCACTAAAGCTTTCAAAGTTTGTAAATAAATTAATAGCATCATTAGCTACATTCATTTTCATATTTATGTGGCATGGAACTGTGTGGAACATATTTGTTTGGTCAAGTCTAAACTACATTGGCATTCTGCTAGAATATACTGGTAAAGCAGTATCAAGTACTGAGGGATACAAATGGTTCAGAACTAAAATTTTAAGAATTCACTAA
Protein
MSETLAIKSLPTLALCGGGLWMGLVFHLKYVISYGTTGAFARLDNMEPPPIPRCVARIHVYSLMWRYFDVGLYQFLSKYIYKPGYSVLSNSLKLSKFVNKLIASLATFIFIFMWHGTVWNIFVWSSLNYIGILLEYTGKAVSSTEGYKWFRTKILRIH
Summary
Similarity
Belongs to the membrane-bound acyltransferase family.
Uniprot
A0A194QYT9
A0A2A4JTP1
A0A194PNM9
A0A2H1VX39
A0A0L7L0U9
A0A1Y1LRD6
+ More
A0A1Y1LPQ2 A0A1B0CD87 A0A1W4WVD4 A0A088AAI9 A0A1D2NBT2 E2B875 A0A2P8Y8C9 A0A1W4X668 A0A1W4X4S9 A0A026WMN8 A0A2A3E2Y4 A0A151IPH2 A0A1B6GCJ1 A0A158NVU2 F4WNR6 A0A067R3L2 A0A1B6MT53 T1IPU2 A0A151XBW9 A0A1B6DAC9 D6WU85 N6UKQ5 A0A1B6IR26 U4U2K1 A0A183J4K0 A0A1B6EDN9 A0A154PCI9 A0A2H8TZR4 A0A2J7QWX9 A0A195FV33 A0A1I8NBE6 A0A0N5E4T7 A0A1J1IET3 A0A336M7A9 A0A195EC48 A0A0V0WWP3 A0A0V0ZKN4 A0A0V0WXF0 A0A0V1CGM9 A0A0V0WWV6 A0A0V0WWM4 F1L3V9 A0A0V0UPQ0 A0A0V1CGR7 A0A0M3HMZ2 A0A0V1LP15 A0A0V0UPH4 A0A1I8P814 A0A0V0UAL4 A0A1I8P841 A0A0V1CHF0 A0A195B7R2 A0A0N4UBN2 E0VJ13 A0A0V1MQY4 A0A0V0RNH6 A0A0C2GJN2 A0A0A9Z0L4 A0A182LTF9 A0A182J2E1 A0A0V1CGW5 A0A0V0UPL8 A0A1Y3ES49 A0A182JYR4 A0A0V1F853 A0A0V1IA08 A0A0V1EAM2 A0A232ERJ1 A0A0B2VFF5 A0A1I8ACR3 A0A183J9J3 A0A0V1HN20 A0A182HSK2 Q7QCF3 A0A182V7D9 A0A182TH46 A0A182Y6I9 A0A182XAR0 A0A182R102 A0A0V0Y4C7 A0A182LEL3 A0A158QXK4 A0A182FT08 W5JTI1 W2SJ96 A0A1A9WFE1 K1PXH5 A0A090L8R5 A0A182PCC9 A0A182W0W5 A0A3P8BBW3 A0A0M4EJF4 A0A077Z2Q2
A0A1Y1LPQ2 A0A1B0CD87 A0A1W4WVD4 A0A088AAI9 A0A1D2NBT2 E2B875 A0A2P8Y8C9 A0A1W4X668 A0A1W4X4S9 A0A026WMN8 A0A2A3E2Y4 A0A151IPH2 A0A1B6GCJ1 A0A158NVU2 F4WNR6 A0A067R3L2 A0A1B6MT53 T1IPU2 A0A151XBW9 A0A1B6DAC9 D6WU85 N6UKQ5 A0A1B6IR26 U4U2K1 A0A183J4K0 A0A1B6EDN9 A0A154PCI9 A0A2H8TZR4 A0A2J7QWX9 A0A195FV33 A0A1I8NBE6 A0A0N5E4T7 A0A1J1IET3 A0A336M7A9 A0A195EC48 A0A0V0WWP3 A0A0V0ZKN4 A0A0V0WXF0 A0A0V1CGM9 A0A0V0WWV6 A0A0V0WWM4 F1L3V9 A0A0V0UPQ0 A0A0V1CGR7 A0A0M3HMZ2 A0A0V1LP15 A0A0V0UPH4 A0A1I8P814 A0A0V0UAL4 A0A1I8P841 A0A0V1CHF0 A0A195B7R2 A0A0N4UBN2 E0VJ13 A0A0V1MQY4 A0A0V0RNH6 A0A0C2GJN2 A0A0A9Z0L4 A0A182LTF9 A0A182J2E1 A0A0V1CGW5 A0A0V0UPL8 A0A1Y3ES49 A0A182JYR4 A0A0V1F853 A0A0V1IA08 A0A0V1EAM2 A0A232ERJ1 A0A0B2VFF5 A0A1I8ACR3 A0A183J9J3 A0A0V1HN20 A0A182HSK2 Q7QCF3 A0A182V7D9 A0A182TH46 A0A182Y6I9 A0A182XAR0 A0A182R102 A0A0V0Y4C7 A0A182LEL3 A0A158QXK4 A0A182FT08 W5JTI1 W2SJ96 A0A1A9WFE1 K1PXH5 A0A090L8R5 A0A182PCC9 A0A182W0W5 A0A3P8BBW3 A0A0M4EJF4 A0A077Z2Q2
Pubmed
EMBL
KQ461150
KPJ08751.1
NWSH01000604
PCG75381.1
KQ459597
KPI94922.1
+ More
ODYU01004977 SOQ45395.1 JTDY01003712 KOB69107.1 GEZM01054521 JAV73587.1 GEZM01054522 JAV73586.1 AJWK01007506 LJIJ01000098 ODN02707.1 GL446286 EFN88077.1 PYGN01000809 PSN40500.1 KK107167 EZA56379.1 KZ288444 PBC25672.1 KQ976892 KYN07238.1 GECZ01009624 JAS60145.1 ADTU01002902 GL888237 EGI64259.1 KK853018 KDR12454.1 GEBQ01000866 JAT39111.1 JH431265 KQ982316 KYQ57855.1 GEDC01014692 JAS22606.1 KQ971352 EFA07646.2 APGK01007092 APGK01029424 KB740734 KB737107 ENN79267.1 ENN83082.1 GECU01018322 JAS89384.1 KB630572 KB632025 ERL83691.1 ERL88104.1 UZAM01014583 VDP34745.1 GEDC01001242 JAS36056.1 KQ434870 KZC09562.1 GFXV01007436 MBW19241.1 NEVH01009412 PNF33093.1 KQ981215 KYN44500.1 CVRI01000048 CRK98783.1 UFQS01000653 UFQT01000653 SSX05797.1 SSX26156.1 KQ979074 KYN22798.1 JYDK01000044 KRX80188.1 JYDQ01000152 KRY12939.1 KRX80190.1 JYDI01000206 KRY48463.1 KRX80189.1 KRX80191.1 JI170854 ADY44813.1 JYDN01000209 KRX53227.1 KRY48464.1 JYDW01000024 KRZ60898.1 KRX53225.1 JYDJ01000037 KRX47863.1 KRY48462.1 KQ976565 KYM80551.1 UYYG01001169 VDN58532.1 DS235219 EEB13369.1 JYDO01000056 KRZ73983.1 JYDL01000116 KRX16048.1 KN732351 KIH59089.1 GBHO01039607 GBHO01006716 JAG03997.1 JAG36888.1 AXCM01000145 KRY48465.1 KRX53226.1 LVZM01003888 OUC47715.1 JYDT01000194 KRY81983.1 JYDS01000271 JYDV01000104 KRZ19667.1 KRZ33383.1 JYDR01000066 KRY70853.1 NNAY01002610 OXU20951.1 JPKZ01001763 KHN80137.1 UZAM01018082 VDP49456.1 JYDP01000043 KRZ12158.1 APCN01000281 AAAB01008859 EAA07632.5 AXCN02001459 JYDU01000065 KRX94866.1 UYSL01019850 VDL70604.1 ADMH02000418 ETN66593.1 KI669059 ETN69724.1 JH816425 EKC21090.1 LN609529 CEF66127.1 UZAH01028792 VDP02394.1 CP012525 ALC45065.1 HG805891 CDW54321.1
ODYU01004977 SOQ45395.1 JTDY01003712 KOB69107.1 GEZM01054521 JAV73587.1 GEZM01054522 JAV73586.1 AJWK01007506 LJIJ01000098 ODN02707.1 GL446286 EFN88077.1 PYGN01000809 PSN40500.1 KK107167 EZA56379.1 KZ288444 PBC25672.1 KQ976892 KYN07238.1 GECZ01009624 JAS60145.1 ADTU01002902 GL888237 EGI64259.1 KK853018 KDR12454.1 GEBQ01000866 JAT39111.1 JH431265 KQ982316 KYQ57855.1 GEDC01014692 JAS22606.1 KQ971352 EFA07646.2 APGK01007092 APGK01029424 KB740734 KB737107 ENN79267.1 ENN83082.1 GECU01018322 JAS89384.1 KB630572 KB632025 ERL83691.1 ERL88104.1 UZAM01014583 VDP34745.1 GEDC01001242 JAS36056.1 KQ434870 KZC09562.1 GFXV01007436 MBW19241.1 NEVH01009412 PNF33093.1 KQ981215 KYN44500.1 CVRI01000048 CRK98783.1 UFQS01000653 UFQT01000653 SSX05797.1 SSX26156.1 KQ979074 KYN22798.1 JYDK01000044 KRX80188.1 JYDQ01000152 KRY12939.1 KRX80190.1 JYDI01000206 KRY48463.1 KRX80189.1 KRX80191.1 JI170854 ADY44813.1 JYDN01000209 KRX53227.1 KRY48464.1 JYDW01000024 KRZ60898.1 KRX53225.1 JYDJ01000037 KRX47863.1 KRY48462.1 KQ976565 KYM80551.1 UYYG01001169 VDN58532.1 DS235219 EEB13369.1 JYDO01000056 KRZ73983.1 JYDL01000116 KRX16048.1 KN732351 KIH59089.1 GBHO01039607 GBHO01006716 JAG03997.1 JAG36888.1 AXCM01000145 KRY48465.1 KRX53226.1 LVZM01003888 OUC47715.1 JYDT01000194 KRY81983.1 JYDS01000271 JYDV01000104 KRZ19667.1 KRZ33383.1 JYDR01000066 KRY70853.1 NNAY01002610 OXU20951.1 JPKZ01001763 KHN80137.1 UZAM01018082 VDP49456.1 JYDP01000043 KRZ12158.1 APCN01000281 AAAB01008859 EAA07632.5 AXCN02001459 JYDU01000065 KRX94866.1 UYSL01019850 VDL70604.1 ADMH02000418 ETN66593.1 KI669059 ETN69724.1 JH816425 EKC21090.1 LN609529 CEF66127.1 UZAH01028792 VDP02394.1 CP012525 ALC45065.1 HG805891 CDW54321.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000037510
UP000092461
UP000192223
+ More
UP000005203 UP000094527 UP000008237 UP000245037 UP000053097 UP000242457 UP000078542 UP000005205 UP000007755 UP000027135 UP000075809 UP000007266 UP000019118 UP000030742 UP000050793 UP000076502 UP000235965 UP000078541 UP000095301 UP000046395 UP000183832 UP000078492 UP000054673 UP000054783 UP000054653 UP000054681 UP000036681 UP000054721 UP000095300 UP000055048 UP000078540 UP000038040 UP000274756 UP000009046 UP000054843 UP000054630 UP000075883 UP000075880 UP000243006 UP000075881 UP000054995 UP000054805 UP000054826 UP000054632 UP000215335 UP000031036 UP000095287 UP000055024 UP000075840 UP000007062 UP000075903 UP000075902 UP000076408 UP000076407 UP000075886 UP000054815 UP000075882 UP000038043 UP000271162 UP000069272 UP000000673 UP000091820 UP000005408 UP000035682 UP000075885 UP000075920 UP000092553 UP000030665
UP000005203 UP000094527 UP000008237 UP000245037 UP000053097 UP000242457 UP000078542 UP000005205 UP000007755 UP000027135 UP000075809 UP000007266 UP000019118 UP000030742 UP000050793 UP000076502 UP000235965 UP000078541 UP000095301 UP000046395 UP000183832 UP000078492 UP000054673 UP000054783 UP000054653 UP000054681 UP000036681 UP000054721 UP000095300 UP000055048 UP000078540 UP000038040 UP000274756 UP000009046 UP000054843 UP000054630 UP000075883 UP000075880 UP000243006 UP000075881 UP000054995 UP000054805 UP000054826 UP000054632 UP000215335 UP000031036 UP000095287 UP000055024 UP000075840 UP000007062 UP000075903 UP000075902 UP000076408 UP000076407 UP000075886 UP000054815 UP000075882 UP000038043 UP000271162 UP000069272 UP000000673 UP000091820 UP000005408 UP000035682 UP000075885 UP000075920 UP000092553 UP000030665
Pfam
Interpro
IPR004299
MBOAT_fam
+ More
IPR032981 HHAT
IPR021133 HEAT_type_2
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR000357 HEAT
IPR005097 Sacchrp_dh_NADP
IPR036291 NAD(P)-bd_dom_sf
IPR009001 Transl_elong_EF1A/Init_IF2_C
IPR015033 HBS1-like_N
IPR000795 TF_GTP-bd_dom
IPR037189 HBS1-like_N_sf
IPR004160 Transl_elong_EFTu/EF1A_C
IPR009000 Transl_B-barrel_sf
IPR027417 P-loop_NTPase
IPR032981 HHAT
IPR021133 HEAT_type_2
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR000357 HEAT
IPR005097 Sacchrp_dh_NADP
IPR036291 NAD(P)-bd_dom_sf
IPR009001 Transl_elong_EF1A/Init_IF2_C
IPR015033 HBS1-like_N
IPR000795 TF_GTP-bd_dom
IPR037189 HBS1-like_N_sf
IPR004160 Transl_elong_EFTu/EF1A_C
IPR009000 Transl_B-barrel_sf
IPR027417 P-loop_NTPase
SUPFAM
Gene 3D
ProteinModelPortal
A0A194QYT9
A0A2A4JTP1
A0A194PNM9
A0A2H1VX39
A0A0L7L0U9
A0A1Y1LRD6
+ More
A0A1Y1LPQ2 A0A1B0CD87 A0A1W4WVD4 A0A088AAI9 A0A1D2NBT2 E2B875 A0A2P8Y8C9 A0A1W4X668 A0A1W4X4S9 A0A026WMN8 A0A2A3E2Y4 A0A151IPH2 A0A1B6GCJ1 A0A158NVU2 F4WNR6 A0A067R3L2 A0A1B6MT53 T1IPU2 A0A151XBW9 A0A1B6DAC9 D6WU85 N6UKQ5 A0A1B6IR26 U4U2K1 A0A183J4K0 A0A1B6EDN9 A0A154PCI9 A0A2H8TZR4 A0A2J7QWX9 A0A195FV33 A0A1I8NBE6 A0A0N5E4T7 A0A1J1IET3 A0A336M7A9 A0A195EC48 A0A0V0WWP3 A0A0V0ZKN4 A0A0V0WXF0 A0A0V1CGM9 A0A0V0WWV6 A0A0V0WWM4 F1L3V9 A0A0V0UPQ0 A0A0V1CGR7 A0A0M3HMZ2 A0A0V1LP15 A0A0V0UPH4 A0A1I8P814 A0A0V0UAL4 A0A1I8P841 A0A0V1CHF0 A0A195B7R2 A0A0N4UBN2 E0VJ13 A0A0V1MQY4 A0A0V0RNH6 A0A0C2GJN2 A0A0A9Z0L4 A0A182LTF9 A0A182J2E1 A0A0V1CGW5 A0A0V0UPL8 A0A1Y3ES49 A0A182JYR4 A0A0V1F853 A0A0V1IA08 A0A0V1EAM2 A0A232ERJ1 A0A0B2VFF5 A0A1I8ACR3 A0A183J9J3 A0A0V1HN20 A0A182HSK2 Q7QCF3 A0A182V7D9 A0A182TH46 A0A182Y6I9 A0A182XAR0 A0A182R102 A0A0V0Y4C7 A0A182LEL3 A0A158QXK4 A0A182FT08 W5JTI1 W2SJ96 A0A1A9WFE1 K1PXH5 A0A090L8R5 A0A182PCC9 A0A182W0W5 A0A3P8BBW3 A0A0M4EJF4 A0A077Z2Q2
A0A1Y1LPQ2 A0A1B0CD87 A0A1W4WVD4 A0A088AAI9 A0A1D2NBT2 E2B875 A0A2P8Y8C9 A0A1W4X668 A0A1W4X4S9 A0A026WMN8 A0A2A3E2Y4 A0A151IPH2 A0A1B6GCJ1 A0A158NVU2 F4WNR6 A0A067R3L2 A0A1B6MT53 T1IPU2 A0A151XBW9 A0A1B6DAC9 D6WU85 N6UKQ5 A0A1B6IR26 U4U2K1 A0A183J4K0 A0A1B6EDN9 A0A154PCI9 A0A2H8TZR4 A0A2J7QWX9 A0A195FV33 A0A1I8NBE6 A0A0N5E4T7 A0A1J1IET3 A0A336M7A9 A0A195EC48 A0A0V0WWP3 A0A0V0ZKN4 A0A0V0WXF0 A0A0V1CGM9 A0A0V0WWV6 A0A0V0WWM4 F1L3V9 A0A0V0UPQ0 A0A0V1CGR7 A0A0M3HMZ2 A0A0V1LP15 A0A0V0UPH4 A0A1I8P814 A0A0V0UAL4 A0A1I8P841 A0A0V1CHF0 A0A195B7R2 A0A0N4UBN2 E0VJ13 A0A0V1MQY4 A0A0V0RNH6 A0A0C2GJN2 A0A0A9Z0L4 A0A182LTF9 A0A182J2E1 A0A0V1CGW5 A0A0V0UPL8 A0A1Y3ES49 A0A182JYR4 A0A0V1F853 A0A0V1IA08 A0A0V1EAM2 A0A232ERJ1 A0A0B2VFF5 A0A1I8ACR3 A0A183J9J3 A0A0V1HN20 A0A182HSK2 Q7QCF3 A0A182V7D9 A0A182TH46 A0A182Y6I9 A0A182XAR0 A0A182R102 A0A0V0Y4C7 A0A182LEL3 A0A158QXK4 A0A182FT08 W5JTI1 W2SJ96 A0A1A9WFE1 K1PXH5 A0A090L8R5 A0A182PCC9 A0A182W0W5 A0A3P8BBW3 A0A0M4EJF4 A0A077Z2Q2
Ontologies
GO
PANTHER
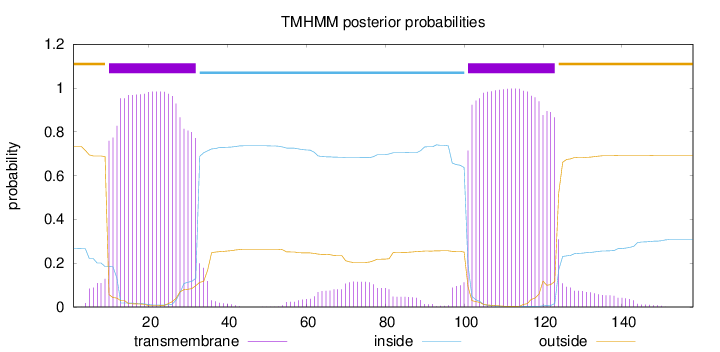
Topology
Length:
158
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
48.28961
Exp number, first 60 AAs:
22.29707
Total prob of N-in:
0.26677
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 100
TMhelix
101 - 123
outside
124 - 158
Population Genetic Test Statistics
Pi
157.744833
Theta
163.03245
Tajima's D
-0.854745
CLR
102.481314
CSRT
0.167241637918104
Interpretation
Uncertain
