Gene
KWMTBOMO02803
Pre Gene Modal
BGIBMGA003507
Annotation
PREDICTED:_DNA_helicase_MCM8_[Amyelois_transitella]
Full name
DNA helicase MCM8
Alternative Name
Minichromosome maintenance 8
Location in the cell
Nuclear Reliability : 2.38
Sequence
CDS
ATGAGACGCACCGGAAGAGGGCGATATCGCAACAACTGGTATCGTTCTAAAAATAAAGGAAACATTAACTCCGATGATTCGAGCCTCAACAACACTTCAACATCCGCAAGGGTCACTAGTACTGCTGGGTCTTCAAGAGCACCAGCTAATCAGCTTCCCATAGTAATATCTGTTGGAACGGATCTTAACAACATATGGAAATTGTACTTTCCGTCCGATGAAAAAGGATCCAATGAAACAGTGGCTAAGCATATTAATAGTTTCAAGCTATATATAAACAAGAATAGGAACTTGTTCGATTTGGAAAAAATAGATCAAACTCACAGCGTACCATTGGATGTGCAGAACTTGCAAAATGATGATGAATTTATTCAAGAGTGGAGTTCCTTTACTACTGACCTTTTTGAGAATCCAGTCACAACATTGTCCACACTAGAATATTGCTTGCATGAGACTTTAAACTGTGCAGTAAAACTAAAAGTGAGAATACTAAATCATCAACCAGTTATACCAATTGCGAGTTTGAAAGTTAATTATTTTGGTAAATTAGTAACAATAAAAGGAACAGTAATAAGAGTGGGCAGTATTGGACTTATTTGCACATCCATGGCGTTCGAATGTTCCAGTTGTCACAGTGTTCAAGCTGTTATTCAGCCACAAGGAGTATTTACTGCTCCGAACTTTTGTCAGAGTTGTAATAGTGGCGTTAAGTTTGAGCCTCTACAGTCCTCGCCGTACACGAACACCACAGATTGGCAGGTTGCAAAAATACAAGAGATCCAGTCTCATAGCATTTCCGGTACTATACCTAGAAGTGTTGAGATTGAGCTCCAAGGAGATTTAGTTGGGACTGCATGTCCAGGAGACGTGCTTTCAGTCACTGGTATAGTACAGGTCCGTGGCGAGAGTAAAGGCGGAGAAGACGGAAAGAGAGCTGCGAGATTGTTGCAGTTATACATTGAAGCTATTTCCATTCACAGTCAGAGGAACTTATCCAATCCGACACTGTCCTTTACTTTAAAGGATTATTATGCAATACAGGAAATTCATGGATCCGACGACGTATTCCGCTTACTAGTTCACTCGCTGTGTCCGACCATCTTTGGTCATGAAGCTGTGAAGGCCGGCCTTATCCTGGGGTTGCTGGGTGGAACTGAGCATGAGAACGGTCCGAGATCTAATCCTCACATACTCGTTGTTGGAGATCCAGGACTGGGCAAATCGCAGTTGTTACAGGCTGCAGCGCATGCTGCGCCCAGGGGTGTGTATGTGTGTGGAGCTTCGGCATCAGCGGGAGGACTGACAGTGGCATTAGGCAGGGAGGCCGGAGGTGATTTCGCGCTCGAAGCTGGCGCATTAGTTTTGGCTGACAAAGGCGTTTGCTGTATTGACGAACTAGATAAAATGAAAGCGCATCACAGTAGTCTGCTGGAGGCCATGGAACAGCGGCGTGTGAGTGTTGCCAAGGGAGGTGTGGTGTGCAGCCTAGCCGCTCGGGCCACAGTGCTTGCCGCCGCAAATCCCGCTGCTGGAAGCTATAATAGAGCAAAAACGGTGGTCGAAAATTTGAAACTAAATTCAGCGCTACTTTCGAGGTTTGATCTAGTTTTTATACTTTTGGATCAACCGGATGAGAAAATGGACGCAATGCTCTCTGAACATGTGTTAGCGCTTCACGCTGGTCATAAAACAAAGAAATCGGCACAGGGTAGCAGTTTTAATAGCAGCATGCATACTAGTCAGAGTAATAATAGCAAACGTTCTCTTAGTCAAAGACTCAGACTGAAGCCGGGAGAGGAGATAGACAATTTACCTCTTGTATTGTTACGGAAGTACATAGCTTACGCACAGAGATATGTACATCCGAAATTGAGTAAAGAAGCGGCAACTGCGCTACAGGAATTTTATCTGGAGCTGCGTCACAACCACGACGCGGATCCTAGTGGTGAACTACGGGAAGAGGCAACAGTGGATGATGCTAATGATGTTATTAGCTTAATGAAACACAGCTTGATCGACACCTTTAGCGATGAATTTGGAAACATTCAACTATCTCGGTCTATCAACGGTTCTGGTGTCAGTTCAAAAAGCAAGTTGAAGAAATATTTGGAGGCATTAACAAGACAATCACATCAGTTAGGAAAAGATATTTTTACACGACAAGAGTTGATACAAATACACAAAGCAGCTGGAGTCCAGGGTGACGCAAATGACCTAATTGAAGCAATGCACATACATTCCTACTTGCTTCTTAAAGCATCTAATACATATCAACTTATACCTATATGA
Protein
MRRTGRGRYRNNWYRSKNKGNINSDDSSLNNTSTSARVTSTAGSSRAPANQLPIVISVGTDLNNIWKLYFPSDEKGSNETVAKHINSFKLYINKNRNLFDLEKIDQTHSVPLDVQNLQNDDEFIQEWSSFTTDLFENPVTTLSTLEYCLHETLNCAVKLKVRILNHQPVIPIASLKVNYFGKLVTIKGTVIRVGSIGLICTSMAFECSSCHSVQAVIQPQGVFTAPNFCQSCNSGVKFEPLQSSPYTNTTDWQVAKIQEIQSHSISGTIPRSVEIELQGDLVGTACPGDVLSVTGIVQVRGESKGGEDGKRAARLLQLYIEAISIHSQRNLSNPTLSFTLKDYYAIQEIHGSDDVFRLLVHSLCPTIFGHEAVKAGLILGLLGGTEHENGPRSNPHILVVGDPGLGKSQLLQAAAHAAPRGVYVCGASASAGGLTVALGREAGGDFALEAGALVLADKGVCCIDELDKMKAHHSSLLEAMEQRRVSVAKGGVVCSLAARATVLAAANPAAGSYNRAKTVVENLKLNSALLSRFDLVFILLDQPDEKMDAMLSEHVLALHAGHKTKKSAQGSSFNSSMHTSQSNNSKRSLSQRLRLKPGEEIDNLPLVLLRKYIAYAQRYVHPKLSKEAATALQEFYLELRHNHDADPSGELREEATVDDANDVISLMKHSLIDTFSDEFGNIQLSRSINGSGVSSKSKLKKYLEALTRQSHQLGKDIFTRQELIQIHKAAGVQGDANDLIEAMHIHSYLLLKASNTYQLIPI
Summary
Description
Component of the MCM8-MCM9 complex, a complex involved in the repair of double-stranded DNA breaks (DBSs) and DNA interstrand cross-links (ICLs) by homologous recombination (HR). Required for DNA resection by the MRE11-RAD50-NBN/NBS1 (MRN) complex by recruiting the MRN complex to the repair site and by promoting the complex nuclease activity. Probably by regulating the localization of the MNR complex, indirectly regulates the recruitment of downstream effector RAD51 to DNA damage sites including DBSs and ICLs. The MCM8-MCM9 complex is dispensable for DNA replication and S phase progression. However, may play a non-essential for DNA replication: may be involved in the activation of the prereplicative complex (pre-RC) during G(1) phase by recruiting CDC6 to the origin recognition complex (ORC). Probably by regulating HR, plays a key role during gametogenesis. Stabilizes MCM9 protein.
Component of the MCM8-MCM9 complex, a complex involved in the repair of double-stranded DNA breaks (DBSs) and DNA interstrand cross-links (ICLs) by homologous recombination (HR). Required for DNA resection by the MRE11-RAD50-NBN/NBS1 (MRN) complex by recruiting the MRN complex to the repair site and by promoting the complex nuclease activity. Probably by regulating the localization of the MNR complex, indirectly regulates the recruitment of downstream effector RAD51 to DNA damage sites including DBSs and ICLs. The MCM8-MCM9 complex is dispensable for DNA replication and S phase progression. However, may play a non-essential for DNA replication: may be involved in the activation of the prereplicative complex (pre-RC) during G(1) phase by recruiting CDC6 to the origin recognition complex (ORC) (By similarity). Probably by regulating HR, plays a key role during gametogenesis (PubMed:22771120). Stabilizes MCM9 protein (By similarity).
Component of the MCM8-MCM9 complex, a complex involved in homologous recombination repair following DNA interstrand cross-links and plays a key role during gametogenesis. The MCM8-MCM9 complex probably acts as a hexameric helicase required to process aberrant forks into homologous recombination substrates and to orchestrate homologous recombination with resection, fork stabilization and fork restart.
Component of the MCM8-MCM9 complex, a complex involved in the repair of double-stranded DNA breaks (DBSs) and DNA interstrand cross-links (ICLs) by homologous recombination (HR) (PubMed:23401855). Required for DNA resection by the MRE11-RAD50-NBN/NBS1 (MRN) complex by recruiting the MRN complex to the repair site and by promoting the complex nuclease activity (PubMed:26215093). Probably by regulating the localization of the MNR complex, indirectly regulates the recruitment of downstream effector RAD51 to DNA damage sites including DBSs and ICLs (PubMed:23401855). The MCM8-MCM9 complex is dispensable for DNA replication and S phase progression (PubMed:23401855). However, may play a non-essential for DNA replication: may be involved in the activation of the prereplicative complex (pre-RC) during G(1) phase by recruiting CDC6 to the origin recognition complex (ORC) (PubMed:15684404). Probably by regulating HR, plays a key role during gametogenesis (By similarity). Stabilizes MCM9 protein (PubMed:23401855, PubMed:26215093).
Component of the MCM8-MCM9 complex, a complex involved in the repair of double-stranded DNA breaks (DBSs) and DNA interstrand cross-links (ICLs) by homologous recombination (HR). Required for DNA resection by the MRE11-RAD50-NBN/NBS1 (MRN) complex by recruiting the MRN complex to the repair site and by promoting the complex nuclease activity. Probably by regulating the localization of the MNR complex, indirectly regulates the recruitment of downstream effector RAD51 to DNA damage sites including DBSs and ICLs. The MCM8-MCM9 complex is dispensable for DNA replication and S phase progression. However, may play a non-essential for DNA replication: may be involved in the activation of the prereplicative complex (pre-RC) during G(1) phase by recruiting CDC6 to the origin recognition complex (ORC) (By similarity). Probably by regulating HR, plays a key role during gametogenesis (PubMed:22771120). Stabilizes MCM9 protein (By similarity).
Component of the MCM8-MCM9 complex, a complex involved in homologous recombination repair following DNA interstrand cross-links and plays a key role during gametogenesis. The MCM8-MCM9 complex probably acts as a hexameric helicase required to process aberrant forks into homologous recombination substrates and to orchestrate homologous recombination with resection, fork stabilization and fork restart.
Component of the MCM8-MCM9 complex, a complex involved in the repair of double-stranded DNA breaks (DBSs) and DNA interstrand cross-links (ICLs) by homologous recombination (HR) (PubMed:23401855). Required for DNA resection by the MRE11-RAD50-NBN/NBS1 (MRN) complex by recruiting the MRN complex to the repair site and by promoting the complex nuclease activity (PubMed:26215093). Probably by regulating the localization of the MNR complex, indirectly regulates the recruitment of downstream effector RAD51 to DNA damage sites including DBSs and ICLs (PubMed:23401855). The MCM8-MCM9 complex is dispensable for DNA replication and S phase progression (PubMed:23401855). However, may play a non-essential for DNA replication: may be involved in the activation of the prereplicative complex (pre-RC) during G(1) phase by recruiting CDC6 to the origin recognition complex (ORC) (PubMed:15684404). Probably by regulating HR, plays a key role during gametogenesis (By similarity). Stabilizes MCM9 protein (PubMed:23401855, PubMed:26215093).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Component of the MCM8-MCM9 complex, which forms a hexamer composed of MCM8 and MCM9. Interacts with the DNA mismatch repair (MMR) complex composed at least of MSH2, MSH3, MSH6, PMS1 and MLH1. Interacts with RAD51; the interaction recruits RAD51 to DNA damage sites. Interacts with the MRN complex composed of MRE11, RAD50 and NBN/NBS1. Interacts with CDC6 and ORC2.
Component of the MCM8-MCM9 complex, which forms a hexamer composed of MCM8 and MCM9.
Component of the MCM8-MCM9 complex, which forms a hexamer composed of MCM8 and MCM9 (PubMed:23401855, PubMed:26300262). Interacts with the DNA mismatch repair (MMR) complex composed at least of MSH2, MSH3, MSH6, PMS1 and MLH1 (PubMed:26300262). Interacts with RAD51; the interaction recruits RAD51 to DNA damage sites (PubMed:23401855). Interacts with the MRN complex composed of MRE11, RAD50 and NBN/NBS1 (PubMed:26215093). Interacts with CDC6 and ORC2 (PubMed:15684404).
Component of the MCM8-MCM9 complex, which forms a hexamer composed of MCM8 and MCM9.
Component of the MCM8-MCM9 complex, which forms a hexamer composed of MCM8 and MCM9 (PubMed:23401855, PubMed:26300262). Interacts with the DNA mismatch repair (MMR) complex composed at least of MSH2, MSH3, MSH6, PMS1 and MLH1 (PubMed:26300262). Interacts with RAD51; the interaction recruits RAD51 to DNA damage sites (PubMed:23401855). Interacts with the MRN complex composed of MRE11, RAD50 and NBN/NBS1 (PubMed:26215093). Interacts with CDC6 and ORC2 (PubMed:15684404).
Similarity
Belongs to the MCM family.
Keywords
ATP-binding
Cell cycle
Chromosome
Complete proteome
DNA damage
DNA repair
DNA replication
DNA-binding
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Alternative splicing
Disease mutation
Polymorphism
Premature ovarian failure
Feature
chain DNA helicase MCM8
splice variant In isoform 2.
sequence variant In dbSNP:rs236110.
splice variant In isoform 2.
sequence variant In dbSNP:rs236110.
Uniprot
H9J1X0
A0A194PNB0
A0A2A4JVE4
A0A2H1VYS5
A0A194QTF9
A0A067RG02
+ More
A0A2P8YK31 A0A2J7QG22 A0A2J7QG23 A0A1W4WYH4 A0A1B6C6A1 A0A1B6LNA8 A0A1S3JYB5 R7VJZ7 A0A023F0L8 A0A210R1Y1 E0VG28 G3GSC8 A0A1U7QLN4 D6WRH7 A0A0A9VT68 D3ZVK1 A0A0L8GIE4 C3Y0U6 E7F643 Q9CWV1 A0A2U3XVQ8 A0A2Y9GZ20 A0A3B4BP21 A0A2U3VSL3 A0A286XPR6 A0A3N0XLN9 A0A2I0LT18 I0IUP3 A0A091HYU3 A0A091DHE0 A0A093CE50 A0A091U8Y2 U3IHH1 A0A093FBH0 Q9CWV1-2 G1N6W2 A0A1V4IGG3 A0A091LTT4 A0A094KNV4 A0A091W700 A0A091E9I8 A0A091PIE1 H0UTZ0 A0A0A0B0E6 R0L7Q9 A0A093J988 A0A091IJN7 H0YVF5 A0A091V8G2 W5K050 A0A091RNG9 F6RN32 A0A2K6RWZ0 A0A2K5XE27 A0A096NVF9 A0A087VNV0 A0A2U3XVR3 A0A091FR56 A0A1S3ARQ8 A0A2R9AVA6 A0A2I3RCL0 A0A091SXC8 W5UD94 A0A0P6J4W3 A0A2U3VSL2 U3KGL8 A0A093P4V9 A0A287D654 A0A091MUU6 A0A2K5VMR7 A0A2K5ELK4 A0A087R8D1 Q9UJA3 A0A3B1IZ11 G3S967 M3Y4M7 G1LVR9 A0A3B4DI58 K7BP55 G1K8S6 H0X833 G3WA38 A0A2Y9IUV5 A0A093SB49 A0A0D9RLZ3 A0A0Q3PJA2 W5Q9U0 A0A3L8SR85 A0A151MGY3 A0A2Y9RIC7 A0A384D3C7 A0A2J8SU62 A0A1U7TXA8 S4RU69
A0A2P8YK31 A0A2J7QG22 A0A2J7QG23 A0A1W4WYH4 A0A1B6C6A1 A0A1B6LNA8 A0A1S3JYB5 R7VJZ7 A0A023F0L8 A0A210R1Y1 E0VG28 G3GSC8 A0A1U7QLN4 D6WRH7 A0A0A9VT68 D3ZVK1 A0A0L8GIE4 C3Y0U6 E7F643 Q9CWV1 A0A2U3XVQ8 A0A2Y9GZ20 A0A3B4BP21 A0A2U3VSL3 A0A286XPR6 A0A3N0XLN9 A0A2I0LT18 I0IUP3 A0A091HYU3 A0A091DHE0 A0A093CE50 A0A091U8Y2 U3IHH1 A0A093FBH0 Q9CWV1-2 G1N6W2 A0A1V4IGG3 A0A091LTT4 A0A094KNV4 A0A091W700 A0A091E9I8 A0A091PIE1 H0UTZ0 A0A0A0B0E6 R0L7Q9 A0A093J988 A0A091IJN7 H0YVF5 A0A091V8G2 W5K050 A0A091RNG9 F6RN32 A0A2K6RWZ0 A0A2K5XE27 A0A096NVF9 A0A087VNV0 A0A2U3XVR3 A0A091FR56 A0A1S3ARQ8 A0A2R9AVA6 A0A2I3RCL0 A0A091SXC8 W5UD94 A0A0P6J4W3 A0A2U3VSL2 U3KGL8 A0A093P4V9 A0A287D654 A0A091MUU6 A0A2K5VMR7 A0A2K5ELK4 A0A087R8D1 Q9UJA3 A0A3B1IZ11 G3S967 M3Y4M7 G1LVR9 A0A3B4DI58 K7BP55 G1K8S6 H0X833 G3WA38 A0A2Y9IUV5 A0A093SB49 A0A0D9RLZ3 A0A0Q3PJA2 W5Q9U0 A0A3L8SR85 A0A151MGY3 A0A2Y9RIC7 A0A384D3C7 A0A2J8SU62 A0A1U7TXA8 S4RU69
EC Number
3.6.4.12
Pubmed
19121390
26354079
24845553
29403074
23254933
25474469
+ More
28812685 20566863 21804562 18362917 19820115 25401762 26823975 15057822 18563158 23594743 16141072 19468303 15489334 22771120 21993624 23371554 22771115 15642098 15592404 23749191 20838655 20360741 25329095 17431167 25319552 25362486 22722832 16136131 23127152 12527764 12771218 14702039 11780052 15684404 16325355 18072282 23401855 23186163 26300262 26215093 25437880 22398555 20010809 21709235 20809919 30282656 22293439
28812685 20566863 21804562 18362917 19820115 25401762 26823975 15057822 18563158 23594743 16141072 19468303 15489334 22771120 21993624 23371554 22771115 15642098 15592404 23749191 20838655 20360741 25329095 17431167 25319552 25362486 22722832 16136131 23127152 12527764 12771218 14702039 11780052 15684404 16325355 18072282 23401855 23186163 26300262 26215093 25437880 22398555 20010809 21709235 20809919 30282656 22293439
EMBL
BABH01007795
BABH01007796
BABH01007797
BABH01007798
KQ459597
KPI94921.1
+ More
NWSH01000604 PCG75380.1 ODYU01004977 SOQ45394.1 KQ461150 KPJ08752.1 KK852659 KDR19137.1 PYGN01000537 PSN44603.1 NEVH01014836 PNF27546.1 PNF27547.1 GEDC01028538 JAS08760.1 GEBQ01014737 JAT25240.1 AMQN01004321 KB293181 ELU16340.1 GBBI01003707 JAC15005.1 NEDP02000845 OWF54901.1 DS235131 EEB12334.1 JH000008 EGV93092.1 KQ971351 EFA06449.1 GBHO01044082 GDHC01004766 JAF99521.1 JAQ13863.1 CH473949 KQ421731 KOF76619.1 GG666479 EEN66079.1 CU929122 FP017149 AK010365 AK133858 AL929562 CH466519 BC046780 BC052070 AAKN02012788 AAKN02012789 RJVU01069573 ROI68668.1 AKCR02000106 PKK20573.1 AB689140 AJ851541 AADN02012184 AADN02012185 KL529797 KFO92403.1 KN124112 KFO22181.1 KL462711 KFV14170.1 KK442053 KFQ70518.1 ADON01070671 KK629635 KFV55042.1 LSYS01009753 OPJ58755.1 KK507393 KFP61837.1 KL263830 KFZ60938.1 KK734780 KFR10875.1 KK718162 KFO54718.1 KK659798 KFQ07425.1 KL873574 KGL99233.1 KB743061 EOB01664.1 KK610049 KFW11565.1 KK500568 KFP08889.1 ABQF01039090 KL410781 KFQ99606.1 KK928228 KFQ43978.1 JSUE03004715 JU477514 AFH34318.1 AHZZ02000595 KL500223 KFO14292.1 KL447392 KFO72985.1 AJFE02073284 AJFE02073285 AJFE02073286 AJFE02073287 AACZ04064981 KK488162 KFQ62635.1 JT412778 AHH39991.1 GEBF01006047 JAN97585.1 AGTO01010954 KL225230 KFW69245.1 AGTP01017661 KK837009 KFP80359.1 AQIA01008254 AQIA01008255 KL226206 KFM09735.1 AJ439063 AY158211 AK027644 AK314654 AL035461 CH471133 BC008830 BC080656 BC101054 BC101055 BC101056 BC101057 CABD030115461 AEYP01029146 AEYP01029147 ACTA01090972 GABC01006556 GABF01006251 GABF01006250 GABE01004056 NBAG03000245 JAA04782.1 JAA15894.1 JAA40683.1 PNI62513.1 PNI62515.1 AAQR03053686 AAQR03053687 AAQR03053688 AAQR03053689 AAQR03053690 AEFK01030381 AEFK01030382 KL670748 KFW80054.1 AQIB01162249 LMAW01002489 KQK81059.1 AMGL01025277 AMGL01025278 AMGL01025279 AMGL01025280 AMGL01025281 AMGL01025282 AMGL01025283 QUSF01000008 RLW06978.1 AKHW03006178 KYO23782.1 NDHI03003544 PNJ24305.1 PNJ24309.1
NWSH01000604 PCG75380.1 ODYU01004977 SOQ45394.1 KQ461150 KPJ08752.1 KK852659 KDR19137.1 PYGN01000537 PSN44603.1 NEVH01014836 PNF27546.1 PNF27547.1 GEDC01028538 JAS08760.1 GEBQ01014737 JAT25240.1 AMQN01004321 KB293181 ELU16340.1 GBBI01003707 JAC15005.1 NEDP02000845 OWF54901.1 DS235131 EEB12334.1 JH000008 EGV93092.1 KQ971351 EFA06449.1 GBHO01044082 GDHC01004766 JAF99521.1 JAQ13863.1 CH473949 KQ421731 KOF76619.1 GG666479 EEN66079.1 CU929122 FP017149 AK010365 AK133858 AL929562 CH466519 BC046780 BC052070 AAKN02012788 AAKN02012789 RJVU01069573 ROI68668.1 AKCR02000106 PKK20573.1 AB689140 AJ851541 AADN02012184 AADN02012185 KL529797 KFO92403.1 KN124112 KFO22181.1 KL462711 KFV14170.1 KK442053 KFQ70518.1 ADON01070671 KK629635 KFV55042.1 LSYS01009753 OPJ58755.1 KK507393 KFP61837.1 KL263830 KFZ60938.1 KK734780 KFR10875.1 KK718162 KFO54718.1 KK659798 KFQ07425.1 KL873574 KGL99233.1 KB743061 EOB01664.1 KK610049 KFW11565.1 KK500568 KFP08889.1 ABQF01039090 KL410781 KFQ99606.1 KK928228 KFQ43978.1 JSUE03004715 JU477514 AFH34318.1 AHZZ02000595 KL500223 KFO14292.1 KL447392 KFO72985.1 AJFE02073284 AJFE02073285 AJFE02073286 AJFE02073287 AACZ04064981 KK488162 KFQ62635.1 JT412778 AHH39991.1 GEBF01006047 JAN97585.1 AGTO01010954 KL225230 KFW69245.1 AGTP01017661 KK837009 KFP80359.1 AQIA01008254 AQIA01008255 KL226206 KFM09735.1 AJ439063 AY158211 AK027644 AK314654 AL035461 CH471133 BC008830 BC080656 BC101054 BC101055 BC101056 BC101057 CABD030115461 AEYP01029146 AEYP01029147 ACTA01090972 GABC01006556 GABF01006251 GABF01006250 GABE01004056 NBAG03000245 JAA04782.1 JAA15894.1 JAA40683.1 PNI62513.1 PNI62515.1 AAQR03053686 AAQR03053687 AAQR03053688 AAQR03053689 AAQR03053690 AEFK01030381 AEFK01030382 KL670748 KFW80054.1 AQIB01162249 LMAW01002489 KQK81059.1 AMGL01025277 AMGL01025278 AMGL01025279 AMGL01025280 AMGL01025281 AMGL01025282 AMGL01025283 QUSF01000008 RLW06978.1 AKHW03006178 KYO23782.1 NDHI03003544 PNJ24305.1 PNJ24309.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000027135
UP000245037
+ More
UP000235965 UP000192223 UP000085678 UP000014760 UP000242188 UP000009046 UP000001075 UP000189706 UP000007266 UP000002494 UP000053454 UP000001554 UP000000437 UP000000589 UP000245341 UP000248481 UP000261440 UP000245340 UP000005447 UP000053872 UP000000539 UP000028990 UP000016666 UP000001645 UP000190648 UP000053605 UP000052976 UP000053858 UP000053119 UP000007754 UP000053283 UP000018467 UP000006718 UP000233200 UP000233140 UP000028761 UP000053760 UP000079721 UP000240080 UP000002277 UP000221080 UP000016665 UP000054081 UP000005215 UP000233100 UP000233020 UP000053286 UP000005640 UP000001519 UP000000715 UP000008912 UP000001646 UP000005225 UP000007648 UP000248482 UP000053258 UP000029965 UP000051836 UP000002356 UP000276834 UP000050525 UP000248480 UP000261680 UP000189704 UP000245300
UP000235965 UP000192223 UP000085678 UP000014760 UP000242188 UP000009046 UP000001075 UP000189706 UP000007266 UP000002494 UP000053454 UP000001554 UP000000437 UP000000589 UP000245341 UP000248481 UP000261440 UP000245340 UP000005447 UP000053872 UP000000539 UP000028990 UP000016666 UP000001645 UP000190648 UP000053605 UP000052976 UP000053858 UP000053119 UP000007754 UP000053283 UP000018467 UP000006718 UP000233200 UP000233140 UP000028761 UP000053760 UP000079721 UP000240080 UP000002277 UP000221080 UP000016665 UP000054081 UP000005215 UP000233100 UP000233020 UP000053286 UP000005640 UP000001519 UP000000715 UP000008912 UP000001646 UP000005225 UP000007648 UP000248482 UP000053258 UP000029965 UP000051836 UP000002356 UP000276834 UP000050525 UP000248480 UP000261680 UP000189704 UP000245300
Interpro
Gene 3D
ProteinModelPortal
H9J1X0
A0A194PNB0
A0A2A4JVE4
A0A2H1VYS5
A0A194QTF9
A0A067RG02
+ More
A0A2P8YK31 A0A2J7QG22 A0A2J7QG23 A0A1W4WYH4 A0A1B6C6A1 A0A1B6LNA8 A0A1S3JYB5 R7VJZ7 A0A023F0L8 A0A210R1Y1 E0VG28 G3GSC8 A0A1U7QLN4 D6WRH7 A0A0A9VT68 D3ZVK1 A0A0L8GIE4 C3Y0U6 E7F643 Q9CWV1 A0A2U3XVQ8 A0A2Y9GZ20 A0A3B4BP21 A0A2U3VSL3 A0A286XPR6 A0A3N0XLN9 A0A2I0LT18 I0IUP3 A0A091HYU3 A0A091DHE0 A0A093CE50 A0A091U8Y2 U3IHH1 A0A093FBH0 Q9CWV1-2 G1N6W2 A0A1V4IGG3 A0A091LTT4 A0A094KNV4 A0A091W700 A0A091E9I8 A0A091PIE1 H0UTZ0 A0A0A0B0E6 R0L7Q9 A0A093J988 A0A091IJN7 H0YVF5 A0A091V8G2 W5K050 A0A091RNG9 F6RN32 A0A2K6RWZ0 A0A2K5XE27 A0A096NVF9 A0A087VNV0 A0A2U3XVR3 A0A091FR56 A0A1S3ARQ8 A0A2R9AVA6 A0A2I3RCL0 A0A091SXC8 W5UD94 A0A0P6J4W3 A0A2U3VSL2 U3KGL8 A0A093P4V9 A0A287D654 A0A091MUU6 A0A2K5VMR7 A0A2K5ELK4 A0A087R8D1 Q9UJA3 A0A3B1IZ11 G3S967 M3Y4M7 G1LVR9 A0A3B4DI58 K7BP55 G1K8S6 H0X833 G3WA38 A0A2Y9IUV5 A0A093SB49 A0A0D9RLZ3 A0A0Q3PJA2 W5Q9U0 A0A3L8SR85 A0A151MGY3 A0A2Y9RIC7 A0A384D3C7 A0A2J8SU62 A0A1U7TXA8 S4RU69
A0A2P8YK31 A0A2J7QG22 A0A2J7QG23 A0A1W4WYH4 A0A1B6C6A1 A0A1B6LNA8 A0A1S3JYB5 R7VJZ7 A0A023F0L8 A0A210R1Y1 E0VG28 G3GSC8 A0A1U7QLN4 D6WRH7 A0A0A9VT68 D3ZVK1 A0A0L8GIE4 C3Y0U6 E7F643 Q9CWV1 A0A2U3XVQ8 A0A2Y9GZ20 A0A3B4BP21 A0A2U3VSL3 A0A286XPR6 A0A3N0XLN9 A0A2I0LT18 I0IUP3 A0A091HYU3 A0A091DHE0 A0A093CE50 A0A091U8Y2 U3IHH1 A0A093FBH0 Q9CWV1-2 G1N6W2 A0A1V4IGG3 A0A091LTT4 A0A094KNV4 A0A091W700 A0A091E9I8 A0A091PIE1 H0UTZ0 A0A0A0B0E6 R0L7Q9 A0A093J988 A0A091IJN7 H0YVF5 A0A091V8G2 W5K050 A0A091RNG9 F6RN32 A0A2K6RWZ0 A0A2K5XE27 A0A096NVF9 A0A087VNV0 A0A2U3XVR3 A0A091FR56 A0A1S3ARQ8 A0A2R9AVA6 A0A2I3RCL0 A0A091SXC8 W5UD94 A0A0P6J4W3 A0A2U3VSL2 U3KGL8 A0A093P4V9 A0A287D654 A0A091MUU6 A0A2K5VMR7 A0A2K5ELK4 A0A087R8D1 Q9UJA3 A0A3B1IZ11 G3S967 M3Y4M7 G1LVR9 A0A3B4DI58 K7BP55 G1K8S6 H0X833 G3WA38 A0A2Y9IUV5 A0A093SB49 A0A0D9RLZ3 A0A0Q3PJA2 W5Q9U0 A0A3L8SR85 A0A151MGY3 A0A2Y9RIC7 A0A384D3C7 A0A2J8SU62 A0A1U7TXA8 S4RU69
PDB
4R7Y
E-value=3.28887e-69,
Score=668
Ontologies
GO
GO:0004521
GO:0006270
GO:0003677
GO:0005524
GO:0004386
GO:0007292
GO:0097362
GO:0048232
GO:0000724
GO:0003697
GO:0005634
GO:0042555
GO:0003688
GO:0005694
GO:0006974
GO:0007049
GO:0005654
GO:0032407
GO:0003682
GO:0032408
GO:0036298
GO:0071168
GO:0032406
GO:0050821
GO:0019899
GO:0000082
GO:0006260
GO:0003777
GO:0007018
GO:0030286
GO:0005216
GO:0045211
GO:0006281
GO:0048384
PANTHER
Topology
Subcellular location
Nucleus
Localizes to nuclear foci. Localizes to double-stranded DNA breaks. Binds chromatin throughout the cell cycle. With evidence from 2 publications.
Chromosome Localizes to nuclear foci. Localizes to double-stranded DNA breaks. Binds chromatin throughout the cell cycle. With evidence from 2 publications.
Chromosome Localizes to nuclear foci. Localizes to double-stranded DNA breaks. Binds chromatin throughout the cell cycle. With evidence from 2 publications.
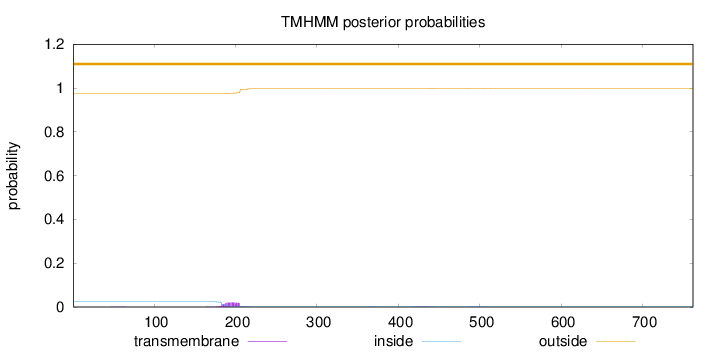
Length:
762
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.516619999999999
Exp number, first 60 AAs:
0.00357
Total prob of N-in:
0.02527
outside
1 - 762
Population Genetic Test Statistics
Pi
281.425899
Theta
169.479811
Tajima's D
1.124668
CLR
0.606404
CSRT
0.696115194240288
Interpretation
Uncertain
