Gene
KWMTBOMO02793 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003694
Annotation
PREDICTED:_sulfotransferase_1C3-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.246
Sequence
CDS
ATGGCTTCTAATAGAAATTGTCCTGAAATACGAGAGAACGGCGAGGAAGAATCGACGGAGCTTCGGAGGCTTTTCAAAGGATTAGAAAACTGCCCTGGATTCGTTCGGGTCGGACCAGCCGGATATTTATTTTCGGTAGCATTTGAACCAGAAATCAAGAGTATATACAACATGGAAGTAAGAGCTACTGATATTTATGTCGCCACATTTCCGAGATCAGGGACAACGTGGACTCAAGAAATGGTCTGGTTACTATTGAATGACTTAGACTACGCCAAGGCGGCCTCACTTAATCTTGTCGACAGATACATAAATGTTGGAATCTCGATCTGGGGTCTAGCATCCAGAGCAAAAGGTGCCAAAATATTAAAATTTAAAGAAGAAGACAAGCGTAAATACAAACAAATAATAACGCCCGGTTGTGAGCTGCTAGCGGCAGCAAGTGACCCTCGCTTCATTAAAACCCATCTTCCAATGTGTATGCTGCCTCCTAATCTCATAGAAACAGCCAAAATAGTATACGTAGCAAGAGATCCACGAGACGTTGTAGTCTCACTGTATCATTTTTACCGTCTTATGCCTGTGCCGCAGTTTACAGGGGACTTCAAAACTTTTTGGAACATGTTCGTCCGCGATAACGTCATCAGAACACCTTATTTCGCACACATAAAAGAGGCATGGGAATTGAGAAATCATCCAAACATGCTATTCCTGTTCTACGAGGAACTTAAAAATGATCTTTCCAATACAATAAAACGCACTGCAAAATTCTTCAACAAAGAATACACAGATGAACAAATCTCAGTGCTGTGCAACCATCTTAATATAAATAATTTCAGACATAACGACTCAGTAAGCTTGAACTTCTTAGCAGATGTGTTTATACCAGGAGAAGAACCATTTATAAGAAAAGGAAAAGTTGGCGGCTGGCGAGATTACTTCGATGACGAGATGACCGTGGAATGCGAACAATGGATGGCCAAAAAACTGAAGGAAACTGGTATTCAGTTCCCGGTTTTCAGTAATGATGTAACCAAGAAATGA
Protein
MASNRNCPEIRENGEEESTELRRLFKGLENCPGFVRVGPAGYLFSVAFEPEIKSIYNMEVRATDIYVATFPRSGTTWTQEMVWLLLNDLDYAKAASLNLVDRYINVGISIWGLASRAKGAKILKFKEEDKRKYKQIITPGCELLAAASDPRFIKTHLPMCMLPPNLIETAKIVYVARDPRDVVVSLYHFYRLMPVPQFTGDFKTFWNMFVRDNVIRTPYFAHIKEAWELRNHPNMLFLFYEELKNDLSNTIKRTAKFFNKEYTDEQISVLCNHLNINNFRHNDSVSLNFLADVFIPGEEPFIRKGKVGGWRDYFDDEMTVECEQWMAKKLKETGIQFPVFSNDVTKK
Summary
Uniprot
H9J2F7
A0A212FII0
H9JB37
A0A2A4KBH0
A0A2H1WDE5
A0A2A4KAH9
+ More
A0A2H1WC50 A0A2H1V382 A0A194QL22 A0A2H1X2F1 A0A2H1WC36 A0A194QI62 A0A0N1IHS9 A0A0N1IET4 A0A194QL64 A0A0N1IJY7 A0A194QHR0 A0A194QHU7 A0A2A4KA51 A0A2H1VZQ2 A0A194QNL9 E2BKD5 E2BKD3 A0A1B0Y0B2 A0A1B6DDS5 A0A194QI57 I4DJT1 A0A194QMT8 K7IXC8 A0A212FIG6 Q26490 E0VIV3 A0A232F572 A0A3Q0JIS4 A0A3Q0JEY6 A0A3Q0JEW1 F4W9H3 A0A212FIH7 A0A2A4KA40 A0A151IWM2 A0A067QQ48 A0A182V8V3 A0A182HKU7 A0A182X9W7 A0A212FIG8 H9JBS7 A0A182N5G4 Q7PXJ0 A0A182L959 A0A2H1VN27 A0A0V0G967 A0A194QJI8 A0A195C9J1 A0A182U3U3 A0PCF9 A0A0L7QLZ9 A0A088AQ03 A0A158N8X8 A0A151WNL9 A0A182YS16 A0A224XS10 A0A182QUI7 A0A0M8ZSK1 E2AHR9 T1IGM8 A0A182P2E0 Q1HPJ5 A0A0V0G778 A0A2A3EI93 A0A224XSY7 A0A023F783 A0A194RJZ8 A0A069DSX1 A0A224XTD2 A0A182RQ05 A0A151HYZ0 E9GE07 A0A194QRV5 A0A069DSB1 A0A084VQC8 A0A182JRW2 A0A2A4K5L1 A0A182JAZ0 A0A195F5F4 A0A0C9RP37 R4WI58 E9IDK2 A0A0L7L630 A0A1L8E4R0 A0A0A9VT74 A0A224XRN0 A0A2J7RQY3 A0A023F7G7 A0A164ZQL2 A0A182M6V5 A0A0P5XSQ1 A0A0P5QLY7 A0A0P5HRC7 A0A0N8AQX5
A0A2H1WC50 A0A2H1V382 A0A194QL22 A0A2H1X2F1 A0A2H1WC36 A0A194QI62 A0A0N1IHS9 A0A0N1IET4 A0A194QL64 A0A0N1IJY7 A0A194QHR0 A0A194QHU7 A0A2A4KA51 A0A2H1VZQ2 A0A194QNL9 E2BKD5 E2BKD3 A0A1B0Y0B2 A0A1B6DDS5 A0A194QI57 I4DJT1 A0A194QMT8 K7IXC8 A0A212FIG6 Q26490 E0VIV3 A0A232F572 A0A3Q0JIS4 A0A3Q0JEY6 A0A3Q0JEW1 F4W9H3 A0A212FIH7 A0A2A4KA40 A0A151IWM2 A0A067QQ48 A0A182V8V3 A0A182HKU7 A0A182X9W7 A0A212FIG8 H9JBS7 A0A182N5G4 Q7PXJ0 A0A182L959 A0A2H1VN27 A0A0V0G967 A0A194QJI8 A0A195C9J1 A0A182U3U3 A0PCF9 A0A0L7QLZ9 A0A088AQ03 A0A158N8X8 A0A151WNL9 A0A182YS16 A0A224XS10 A0A182QUI7 A0A0M8ZSK1 E2AHR9 T1IGM8 A0A182P2E0 Q1HPJ5 A0A0V0G778 A0A2A3EI93 A0A224XSY7 A0A023F783 A0A194RJZ8 A0A069DSX1 A0A224XTD2 A0A182RQ05 A0A151HYZ0 E9GE07 A0A194QRV5 A0A069DSB1 A0A084VQC8 A0A182JRW2 A0A2A4K5L1 A0A182JAZ0 A0A195F5F4 A0A0C9RP37 R4WI58 E9IDK2 A0A0L7L630 A0A1L8E4R0 A0A0A9VT74 A0A224XRN0 A0A2J7RQY3 A0A023F7G7 A0A164ZQL2 A0A182M6V5 A0A0P5XSQ1 A0A0P5QLY7 A0A0P5HRC7 A0A0N8AQX5
Pubmed
EMBL
BABH01007897
AGBW02008382
OWR53534.1
BABH01008529
BABH01008530
BABH01008531
+ More
NWSH01000001 PCG81090.1 ODYU01007908 SOQ51100.1 PCG81089.1 ODYU01007663 SOQ50640.1 ODYU01000444 SOQ35226.1 KQ461198 KPJ06273.1 ODYU01012890 SOQ59422.1 SOQ50641.1 KQ458793 KPJ05084.1 KQ460458 KPJ14654.1 LADJ01060126 KPJ21606.1 KPJ06272.1 KPJ21605.1 KPJ05083.1 KPJ05082.1 PCG81087.1 ODYU01005448 SOQ46321.1 KPJ05081.1 GL448783 EFN83859.1 EFN83857.1 KT000403 ANI86001.1 GEDC01013553 JAS23745.1 KPJ05079.1 AK401549 BAM18171.1 KPJ06275.1 AAZX01000018 OWR53535.1 U28654 AAC47136.1 DS235206 EEB13309.1 NNAY01000980 OXU25629.1 GL888030 EGI69148.1 OWR53536.1 PCG81091.1 KQ980849 KYN12229.1 KK853064 KDR11875.1 APCN01000869 OWR53533.1 BABH01015119 BABH01015120 AAAB01008987 EAA01764.4 ODYU01003446 SOQ42235.1 GECL01001637 JAP04487.1 KPJ05080.1 KQ978068 KYM97385.1 AB284983 BAF37537.1 KQ414905 KOC59625.1 ADTU01008904 ADTU01008905 KQ982907 KYQ49408.1 GFTR01005206 JAW11220.1 AXCN02001171 KQ435922 KOX68659.1 GL439579 EFN66998.1 ACPB03012364 DQ443407 ABF51496.1 GECL01002154 JAP03970.1 KZ288232 PBC31513.1 GFTR01005203 JAW11223.1 GBBI01001612 JAC17100.1 KQ460124 KPJ17660.1 GBGD01002073 JAC86816.1 GFTR01005205 JAW11221.1 KQ976714 KYM76939.1 GL732540 EFX82399.1 KPJ06271.1 GBGD01002074 JAC86815.1 ATLV01015189 KE525003 KFB40172.1 NWSH01000133 PCG79208.1 KQ981810 KYN35407.1 GBYB01015302 JAG85069.1 AK417030 BAN20245.1 GL762454 EFZ21395.1 JTDY01002690 KOB70912.1 GFDF01000367 JAV13717.1 GBHO01044077 GBRD01004179 GDHC01002349 JAF99526.1 JAG61642.1 JAQ16280.1 GFTR01005326 JAW11100.1 NEVH01000613 PNF43242.1 GBBI01001568 JAC17144.1 LRGB01000687 KZS16625.1 AXCM01017667 GDIP01068109 JAM35606.1 GDIQ01112393 JAL39333.1 GDIQ01223415 JAK28310.1 GDIQ01251659 JAK00066.1
NWSH01000001 PCG81090.1 ODYU01007908 SOQ51100.1 PCG81089.1 ODYU01007663 SOQ50640.1 ODYU01000444 SOQ35226.1 KQ461198 KPJ06273.1 ODYU01012890 SOQ59422.1 SOQ50641.1 KQ458793 KPJ05084.1 KQ460458 KPJ14654.1 LADJ01060126 KPJ21606.1 KPJ06272.1 KPJ21605.1 KPJ05083.1 KPJ05082.1 PCG81087.1 ODYU01005448 SOQ46321.1 KPJ05081.1 GL448783 EFN83859.1 EFN83857.1 KT000403 ANI86001.1 GEDC01013553 JAS23745.1 KPJ05079.1 AK401549 BAM18171.1 KPJ06275.1 AAZX01000018 OWR53535.1 U28654 AAC47136.1 DS235206 EEB13309.1 NNAY01000980 OXU25629.1 GL888030 EGI69148.1 OWR53536.1 PCG81091.1 KQ980849 KYN12229.1 KK853064 KDR11875.1 APCN01000869 OWR53533.1 BABH01015119 BABH01015120 AAAB01008987 EAA01764.4 ODYU01003446 SOQ42235.1 GECL01001637 JAP04487.1 KPJ05080.1 KQ978068 KYM97385.1 AB284983 BAF37537.1 KQ414905 KOC59625.1 ADTU01008904 ADTU01008905 KQ982907 KYQ49408.1 GFTR01005206 JAW11220.1 AXCN02001171 KQ435922 KOX68659.1 GL439579 EFN66998.1 ACPB03012364 DQ443407 ABF51496.1 GECL01002154 JAP03970.1 KZ288232 PBC31513.1 GFTR01005203 JAW11223.1 GBBI01001612 JAC17100.1 KQ460124 KPJ17660.1 GBGD01002073 JAC86816.1 GFTR01005205 JAW11221.1 KQ976714 KYM76939.1 GL732540 EFX82399.1 KPJ06271.1 GBGD01002074 JAC86815.1 ATLV01015189 KE525003 KFB40172.1 NWSH01000133 PCG79208.1 KQ981810 KYN35407.1 GBYB01015302 JAG85069.1 AK417030 BAN20245.1 GL762454 EFZ21395.1 JTDY01002690 KOB70912.1 GFDF01000367 JAV13717.1 GBHO01044077 GBRD01004179 GDHC01002349 JAF99526.1 JAG61642.1 JAQ16280.1 GFTR01005326 JAW11100.1 NEVH01000613 PNF43242.1 GBBI01001568 JAC17144.1 LRGB01000687 KZS16625.1 AXCM01017667 GDIP01068109 JAM35606.1 GDIQ01112393 JAL39333.1 GDIQ01223415 JAK28310.1 GDIQ01251659 JAK00066.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000008237
+ More
UP000002358 UP000009046 UP000215335 UP000079169 UP000007755 UP000078492 UP000027135 UP000075903 UP000075840 UP000076407 UP000075884 UP000007062 UP000075882 UP000078542 UP000075902 UP000053825 UP000005203 UP000005205 UP000075809 UP000076408 UP000075886 UP000053105 UP000000311 UP000015103 UP000075885 UP000242457 UP000075900 UP000078540 UP000000305 UP000030765 UP000075881 UP000075880 UP000078541 UP000037510 UP000235965 UP000076858 UP000075883
UP000002358 UP000009046 UP000215335 UP000079169 UP000007755 UP000078492 UP000027135 UP000075903 UP000075840 UP000076407 UP000075884 UP000007062 UP000075882 UP000078542 UP000075902 UP000053825 UP000005203 UP000005205 UP000075809 UP000076408 UP000075886 UP000053105 UP000000311 UP000015103 UP000075885 UP000242457 UP000075900 UP000078540 UP000000305 UP000030765 UP000075881 UP000075880 UP000078541 UP000037510 UP000235965 UP000076858 UP000075883
Interpro
Gene 3D
ProteinModelPortal
H9J2F7
A0A212FII0
H9JB37
A0A2A4KBH0
A0A2H1WDE5
A0A2A4KAH9
+ More
A0A2H1WC50 A0A2H1V382 A0A194QL22 A0A2H1X2F1 A0A2H1WC36 A0A194QI62 A0A0N1IHS9 A0A0N1IET4 A0A194QL64 A0A0N1IJY7 A0A194QHR0 A0A194QHU7 A0A2A4KA51 A0A2H1VZQ2 A0A194QNL9 E2BKD5 E2BKD3 A0A1B0Y0B2 A0A1B6DDS5 A0A194QI57 I4DJT1 A0A194QMT8 K7IXC8 A0A212FIG6 Q26490 E0VIV3 A0A232F572 A0A3Q0JIS4 A0A3Q0JEY6 A0A3Q0JEW1 F4W9H3 A0A212FIH7 A0A2A4KA40 A0A151IWM2 A0A067QQ48 A0A182V8V3 A0A182HKU7 A0A182X9W7 A0A212FIG8 H9JBS7 A0A182N5G4 Q7PXJ0 A0A182L959 A0A2H1VN27 A0A0V0G967 A0A194QJI8 A0A195C9J1 A0A182U3U3 A0PCF9 A0A0L7QLZ9 A0A088AQ03 A0A158N8X8 A0A151WNL9 A0A182YS16 A0A224XS10 A0A182QUI7 A0A0M8ZSK1 E2AHR9 T1IGM8 A0A182P2E0 Q1HPJ5 A0A0V0G778 A0A2A3EI93 A0A224XSY7 A0A023F783 A0A194RJZ8 A0A069DSX1 A0A224XTD2 A0A182RQ05 A0A151HYZ0 E9GE07 A0A194QRV5 A0A069DSB1 A0A084VQC8 A0A182JRW2 A0A2A4K5L1 A0A182JAZ0 A0A195F5F4 A0A0C9RP37 R4WI58 E9IDK2 A0A0L7L630 A0A1L8E4R0 A0A0A9VT74 A0A224XRN0 A0A2J7RQY3 A0A023F7G7 A0A164ZQL2 A0A182M6V5 A0A0P5XSQ1 A0A0P5QLY7 A0A0P5HRC7 A0A0N8AQX5
A0A2H1WC50 A0A2H1V382 A0A194QL22 A0A2H1X2F1 A0A2H1WC36 A0A194QI62 A0A0N1IHS9 A0A0N1IET4 A0A194QL64 A0A0N1IJY7 A0A194QHR0 A0A194QHU7 A0A2A4KA51 A0A2H1VZQ2 A0A194QNL9 E2BKD5 E2BKD3 A0A1B0Y0B2 A0A1B6DDS5 A0A194QI57 I4DJT1 A0A194QMT8 K7IXC8 A0A212FIG6 Q26490 E0VIV3 A0A232F572 A0A3Q0JIS4 A0A3Q0JEY6 A0A3Q0JEW1 F4W9H3 A0A212FIH7 A0A2A4KA40 A0A151IWM2 A0A067QQ48 A0A182V8V3 A0A182HKU7 A0A182X9W7 A0A212FIG8 H9JBS7 A0A182N5G4 Q7PXJ0 A0A182L959 A0A2H1VN27 A0A0V0G967 A0A194QJI8 A0A195C9J1 A0A182U3U3 A0PCF9 A0A0L7QLZ9 A0A088AQ03 A0A158N8X8 A0A151WNL9 A0A182YS16 A0A224XS10 A0A182QUI7 A0A0M8ZSK1 E2AHR9 T1IGM8 A0A182P2E0 Q1HPJ5 A0A0V0G778 A0A2A3EI93 A0A224XSY7 A0A023F783 A0A194RJZ8 A0A069DSX1 A0A224XTD2 A0A182RQ05 A0A151HYZ0 E9GE07 A0A194QRV5 A0A069DSB1 A0A084VQC8 A0A182JRW2 A0A2A4K5L1 A0A182JAZ0 A0A195F5F4 A0A0C9RP37 R4WI58 E9IDK2 A0A0L7L630 A0A1L8E4R0 A0A0A9VT74 A0A224XRN0 A0A2J7RQY3 A0A023F7G7 A0A164ZQL2 A0A182M6V5 A0A0P5XSQ1 A0A0P5QLY7 A0A0P5HRC7 A0A0N8AQX5
PDB
1FML
E-value=7.40713e-71,
Score=678
Ontologies
GO
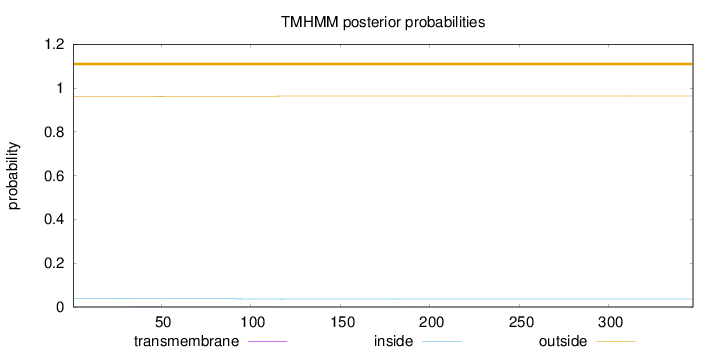
Topology
Length:
347
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0784299999999997
Exp number, first 60 AAs:
0.04595
Total prob of N-in:
0.03946
outside
1 - 347
Population Genetic Test Statistics
Pi
215.53938
Theta
178.31918
Tajima's D
0.229916
CLR
0.275212
CSRT
0.43712814359282
Interpretation
Uncertain
