Pre Gene Modal
BGIBMGA003497
Annotation
uncharacterized_protein_LOC101741261_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.943
Sequence
CDS
ATGAGTGGAAAATATGGAGCGACGGATGTTAATGGAACAACCTGTGATATTCAAGAAGAAATCGTCGTAGATAATGAAGTTGATTCGAGTGCATCGTACAAGAGAAGATGGTGGCGTGTCAGTCAGAACGTGCTTTACGGACTAGGGCATGTTTACAACGACCTCTGCGCTGCGATGTGGTTCTCATACATGCTGCTTTTCTTTCAAGCAGTCCTGGACATGAGAGCTGTCGTAGCTGGAGCTATGCTGCTTCTAGGACAAGTTGTAGATGCACTTGCAACCCCTGTTGTGGGAGTTTTGGCTGATAAATACGGAACTAAGAAGGCATGGCATTTGACTGGATGCATTTTAGTAACAGCGACATTCCCGTTGCTGTTCATTAGATGTTGGGGTTGCTGGTTTAATGAAAACACACAGTATTTGTACTGGTGGATGCCACTATATTATGCGGTCCTAATTATATTTTTCCAAATTGGTTGGGCCGTGGTACAAATCTCACATCTGGCTATGATTCCGTCAATCACCGACAACCTTCAAGTTCGTGCCGAATTGACTTCTATTAGGTATATGGCTTCTGTGATGTCGAGTCTCACTGTTTACCTGATAACTTGGGTTGTTTTGAGGGCAACAACCTACAGTACATTCATCAGCCAAACAGATGATTACAAATTTCGGGACGTCTCCTTGATAATTTCTGGTTTGGGAGTAATATCTTTCATGGCCTTTCATTTATTTTTCAAACTTCGAAGAGAAGATAAATCTGGCAAAGGAGCACCCGTTGTCCAGAACGGTAATGGAACTTTCGTCCACCAAGACAACGAAGCAGCGAAATTACCCGCTAAATCTAAAATTATGTACTTCCTGCGAATGCCGTTGTTATATCAGACCAGTTTATTATATGTATTTTCTCGTCTGTATTGGGCATTGAGTCTTGTGTACGTGCCATTGTTCTTGGAGGAACGTTTATCTGTGAACCCGAGTGCAGGATCAGAGCTAGTTGCGAGTGTACCTCTTGTGCTTTACATTTCATCGTTCTTTTTCTCGCTACTACTCAAGAGCAAGATTAACAGTTTTGGCAACCAGGTGGTGTATTTCATTGGGAGTTGCTTAAGCTTAGGAAGCTGCCTGTGGATTGCATTAGCCATAGATCCGGAAGCGAGCATAGTTCAAATCTACATGGTCGCAACACTCATTGGGGCAGGAAGCTCAATCACGCTCGTCTCCAGCCTATGTGTCACAGCAGATCTCATAGGGCCGCACTCACACCAAAGTGCTGCCATCTATTCTATAGTGACATTTGCAGACAAACTTGTTACGGGTATAGCTGTCGTCGCTATTCAGAACTATAAATGCGATGATGTTGTGGATTGTCCTCAATATTATCGAGGGGTGCTCACGTACGCATGCGGCGGAAGTGCCGCACTTGGCATATTGTCTTTGTCTCTCACTAAACTAATTTCAATCAAAACTAAACCAGAGGTGTAA
Protein
MSGKYGATDVNGTTCDIQEEIVVDNEVDSSASYKRRWWRVSQNVLYGLGHVYNDLCAAMWFSYMLLFFQAVLDMRAVVAGAMLLLGQVVDALATPVVGVLADKYGTKKAWHLTGCILVTATFPLLFIRCWGCWFNENTQYLYWWMPLYYAVLIIFFQIGWAVVQISHLAMIPSITDNLQVRAELTSIRYMASVMSSLTVYLITWVVLRATTYSTFISQTDDYKFRDVSLIISGLGVISFMAFHLFFKLRREDKSGKGAPVVQNGNGTFVHQDNEAAKLPAKSKIMYFLRMPLLYQTSLLYVFSRLYWALSLVYVPLFLEERLSVNPSAGSELVASVPLVLYISSFFFSLLLKSKINSFGNQVVYFIGSCLSLGSCLWIALAIDPEASIVQIYMVATLIGAGSSITLVSSLCVTADLIGPHSHQSAAIYSIVTFADKLVTGIAVVAIQNYKCDDVVDCPQYYRGVLTYACGGSAALGILSLSLTKLISIKTKPEV
Summary
Uniprot
H9J1W0
I0IYT1
A0A2A4JX89
A0A2H1VTG5
A0A212EHK6
A0A194PQ93
+ More
A0A0L7LH67 A0A084WB96 W5JI80 A0A182IT02 A0A2M4AFX5 A0A182V1J7 A0A182LPD5 A0A182HNH1 Q7Q355 A0A2M4BIQ3 A0A2P8ZLX4 A0A182WF88 A0A2M4BIS1 A0A182Q0R5 B0WBU6 A0A182T887 A0A182U5J1 A0A182YJZ9 A0A182PVD0 U5EYC5 A0A182MP96 A0A1L8DE05 A0A182FU18 A0A1B0CW50 E2A529 A0A310SVZ3 I0IYT2 A0A1B6FLL6 A0A1B6F907 D6W6W0 A0A1L8DEZ9 A0A088AUM5 A0A195CQH4 A0A1B6HR18 A0A1W4XGQ0 A0A1Y1LPH1 A0A151X3T6 A0A1W4XFI0 A0A1W4XGF1 A0A151IYH3 A0A1B6LGX7 A0A2J7PGJ2 A0A1B6DK64 A0A1W4XGQ4 A0A1B6L6U8 A0A158NKW0 A0A067R5D6 A0A151JYR5 K7IS83 A0A0L7R7S5 E0VQM9 A0A232ERE3 F4WE57 A0A088AJC7 A0A0L7RBV1 A0A1Y1LD93 E2AJ53 D6W847 A0A139WPG3 A0A0C9QWP9 A0A0P4WDL5 A0A151JVP9 A0A195BKU0 A0A158P455 K7IZF0 A0A195CNW3 A0A151J9B2 A0A2H1VA33 A0A151I5F2 A0A1W0WRE6 A0A1W4XMR4 A0A1W4XDI7 A0A1J1ICW3 A0A087V0H2 E9FVJ5 A0A2A4JTR3 E2BGG8 U4U133 N6TV38 T1JDD2 A0A1B6E3G3 A0A1B6DY20 A0A2W1CHU9 A0A151X0H6 E0VGF6 A0A0P5A7W7 A0A0P5JVG7 A0A0P5AWT0 A0A0P5VXK5 A0A0P5N487 A0A0P4Z4M9 A0A0P5NTE2 A0A0N8A344
A0A0L7LH67 A0A084WB96 W5JI80 A0A182IT02 A0A2M4AFX5 A0A182V1J7 A0A182LPD5 A0A182HNH1 Q7Q355 A0A2M4BIQ3 A0A2P8ZLX4 A0A182WF88 A0A2M4BIS1 A0A182Q0R5 B0WBU6 A0A182T887 A0A182U5J1 A0A182YJZ9 A0A182PVD0 U5EYC5 A0A182MP96 A0A1L8DE05 A0A182FU18 A0A1B0CW50 E2A529 A0A310SVZ3 I0IYT2 A0A1B6FLL6 A0A1B6F907 D6W6W0 A0A1L8DEZ9 A0A088AUM5 A0A195CQH4 A0A1B6HR18 A0A1W4XGQ0 A0A1Y1LPH1 A0A151X3T6 A0A1W4XFI0 A0A1W4XGF1 A0A151IYH3 A0A1B6LGX7 A0A2J7PGJ2 A0A1B6DK64 A0A1W4XGQ4 A0A1B6L6U8 A0A158NKW0 A0A067R5D6 A0A151JYR5 K7IS83 A0A0L7R7S5 E0VQM9 A0A232ERE3 F4WE57 A0A088AJC7 A0A0L7RBV1 A0A1Y1LD93 E2AJ53 D6W847 A0A139WPG3 A0A0C9QWP9 A0A0P4WDL5 A0A151JVP9 A0A195BKU0 A0A158P455 K7IZF0 A0A195CNW3 A0A151J9B2 A0A2H1VA33 A0A151I5F2 A0A1W0WRE6 A0A1W4XMR4 A0A1W4XDI7 A0A1J1ICW3 A0A087V0H2 E9FVJ5 A0A2A4JTR3 E2BGG8 U4U133 N6TV38 T1JDD2 A0A1B6E3G3 A0A1B6DY20 A0A2W1CHU9 A0A151X0H6 E0VGF6 A0A0P5A7W7 A0A0P5JVG7 A0A0P5AWT0 A0A0P5VXK5 A0A0P5N487 A0A0P4Z4M9 A0A0P5NTE2 A0A0N8A344
Pubmed
EMBL
BABH01007901
BABH01007902
BABH01007903
BABH01007904
AB663084
BAM10430.1
+ More
NWSH01000438 PCG76429.1 ODYU01004352 SOQ44141.1 AGBW02014874 OWR40964.1 KQ459597 KPI94909.1 JTDY01001159 KOB74709.1 ATLV01022327 KE525331 KFB47490.1 ADMH02001328 ETN62993.1 GGFK01006362 MBW39683.1 APCN01001930 AAAB01008966 EAA12972.4 GGFJ01003794 MBW52935.1 PYGN01000020 PSN57470.1 GGFJ01003791 MBW52932.1 AXCN02001640 DS231882 EDS42876.1 GANO01000382 JAB59489.1 AXCM01000360 GFDF01009406 JAV04678.1 AJWK01031762 GL436794 EFN71460.1 KQ759862 OAD62523.1 AB663085 BAM10431.1 GECZ01018671 JAS51098.1 GECZ01023064 JAS46705.1 KQ971307 EFA11449.2 GFDF01009127 JAV04957.1 KQ977481 KYN02364.1 GECU01030588 JAS77118.1 GEZM01051757 GEZM01051756 JAV74718.1 KQ982557 KYQ55073.1 KQ980763 KYN13504.1 GEBQ01017089 JAT22888.1 NEVH01025212 PNF15452.1 GEDC01011214 JAS26084.1 GEBQ01020581 JAT19396.1 ADTU01000017 KK852954 KDR13311.1 KQ981463 KYN41517.1 KQ414638 KOC66878.1 DS235430 EEB15685.1 NNAY01002629 OXU20901.1 GL888102 EGI67495.1 KQ414617 KOC68329.1 GEZM01058973 JAV71594.1 GL439967 EFN66493.1 EFA10999.1 KYB29834.1 GBYB01008159 JAG77926.1 GDRN01071210 GDRN01071209 JAI63734.1 KQ981701 KYN37495.1 KQ976453 KYM85340.1 ADTU01008610 KQ977513 KYN02167.1 KQ979480 KYN21281.1 ODYU01001466 SOQ37698.1 KQ976425 KYM88101.1 MTYJ01000056 OQV17781.1 CVRI01000043 CRK96281.1 KK122592 KFM83111.1 GL732525 EFX89093.1 NWSH01000598 PCG75425.1 GL448181 EFN85217.1 KB631768 ERL85998.1 APGK01050965 APGK01050966 KB741180 ENN73125.1 JH432104 GEDC01004847 JAS32451.1 GEDC01006737 JAS30561.1 KZ150098 PZC73585.1 KQ982612 KYQ53868.1 DS235140 EEB12462.1 GDIP01207236 JAJ16166.1 GDIQ01193458 JAK58267.1 GDIP01207237 JAJ16165.1 GDIP01094649 JAM09066.1 GDIQ01149839 JAL01887.1 GDIP01222376 JAJ01026.1 GDIQ01148232 JAL03494.1 GDIP01187185 GDIP01108483 LRGB01002864 JAJ36217.1 KZS05761.1
NWSH01000438 PCG76429.1 ODYU01004352 SOQ44141.1 AGBW02014874 OWR40964.1 KQ459597 KPI94909.1 JTDY01001159 KOB74709.1 ATLV01022327 KE525331 KFB47490.1 ADMH02001328 ETN62993.1 GGFK01006362 MBW39683.1 APCN01001930 AAAB01008966 EAA12972.4 GGFJ01003794 MBW52935.1 PYGN01000020 PSN57470.1 GGFJ01003791 MBW52932.1 AXCN02001640 DS231882 EDS42876.1 GANO01000382 JAB59489.1 AXCM01000360 GFDF01009406 JAV04678.1 AJWK01031762 GL436794 EFN71460.1 KQ759862 OAD62523.1 AB663085 BAM10431.1 GECZ01018671 JAS51098.1 GECZ01023064 JAS46705.1 KQ971307 EFA11449.2 GFDF01009127 JAV04957.1 KQ977481 KYN02364.1 GECU01030588 JAS77118.1 GEZM01051757 GEZM01051756 JAV74718.1 KQ982557 KYQ55073.1 KQ980763 KYN13504.1 GEBQ01017089 JAT22888.1 NEVH01025212 PNF15452.1 GEDC01011214 JAS26084.1 GEBQ01020581 JAT19396.1 ADTU01000017 KK852954 KDR13311.1 KQ981463 KYN41517.1 KQ414638 KOC66878.1 DS235430 EEB15685.1 NNAY01002629 OXU20901.1 GL888102 EGI67495.1 KQ414617 KOC68329.1 GEZM01058973 JAV71594.1 GL439967 EFN66493.1 EFA10999.1 KYB29834.1 GBYB01008159 JAG77926.1 GDRN01071210 GDRN01071209 JAI63734.1 KQ981701 KYN37495.1 KQ976453 KYM85340.1 ADTU01008610 KQ977513 KYN02167.1 KQ979480 KYN21281.1 ODYU01001466 SOQ37698.1 KQ976425 KYM88101.1 MTYJ01000056 OQV17781.1 CVRI01000043 CRK96281.1 KK122592 KFM83111.1 GL732525 EFX89093.1 NWSH01000598 PCG75425.1 GL448181 EFN85217.1 KB631768 ERL85998.1 APGK01050965 APGK01050966 KB741180 ENN73125.1 JH432104 GEDC01004847 JAS32451.1 GEDC01006737 JAS30561.1 KZ150098 PZC73585.1 KQ982612 KYQ53868.1 DS235140 EEB12462.1 GDIP01207236 JAJ16166.1 GDIQ01193458 JAK58267.1 GDIP01207237 JAJ16165.1 GDIP01094649 JAM09066.1 GDIQ01149839 JAL01887.1 GDIP01222376 JAJ01026.1 GDIQ01148232 JAL03494.1 GDIP01187185 GDIP01108483 LRGB01002864 JAJ36217.1 KZS05761.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000037510
UP000030765
+ More
UP000000673 UP000075880 UP000075903 UP000075882 UP000075840 UP000007062 UP000245037 UP000075920 UP000075886 UP000002320 UP000075901 UP000075902 UP000076408 UP000075885 UP000075883 UP000069272 UP000092461 UP000000311 UP000007266 UP000005203 UP000078542 UP000192223 UP000075809 UP000078492 UP000235965 UP000005205 UP000027135 UP000078541 UP000002358 UP000053825 UP000009046 UP000215335 UP000007755 UP000078540 UP000183832 UP000054359 UP000000305 UP000008237 UP000030742 UP000019118 UP000076858
UP000000673 UP000075880 UP000075903 UP000075882 UP000075840 UP000007062 UP000245037 UP000075920 UP000075886 UP000002320 UP000075901 UP000075902 UP000076408 UP000075885 UP000075883 UP000069272 UP000092461 UP000000311 UP000007266 UP000005203 UP000078542 UP000192223 UP000075809 UP000078492 UP000235965 UP000005205 UP000027135 UP000078541 UP000002358 UP000053825 UP000009046 UP000215335 UP000007755 UP000078540 UP000183832 UP000054359 UP000000305 UP000008237 UP000030742 UP000019118 UP000076858
Pfam
Interpro
IPR000330
SNF2_N
+ More
IPR016197 Chromo-like_dom_sf
IPR036259 MFS_trans_sf
IPR023779 Chromodomain_CS
IPR023780 Chromo_domain
IPR038718 SNF2-like_sf
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR020846 MFS_dom
IPR027417 P-loop_NTPase
IPR025260 DUF4208
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR000953 Chromo/chromo_shadow_dom
IPR040793 CDH1_2_SANT_HL1
IPR039672 MFS_2
IPR011701 MFS
IPR036871 PX_dom_sf
IPR001683 Phox
IPR040842 SNX17/31_FERM
IPR011993 PH-like_dom_sf
IPR037836 SNX17_FERM-like_dom
IPR016197 Chromo-like_dom_sf
IPR036259 MFS_trans_sf
IPR023779 Chromodomain_CS
IPR023780 Chromo_domain
IPR038718 SNF2-like_sf
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR020846 MFS_dom
IPR027417 P-loop_NTPase
IPR025260 DUF4208
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR000953 Chromo/chromo_shadow_dom
IPR040793 CDH1_2_SANT_HL1
IPR039672 MFS_2
IPR011701 MFS
IPR036871 PX_dom_sf
IPR001683 Phox
IPR040842 SNX17/31_FERM
IPR011993 PH-like_dom_sf
IPR037836 SNX17_FERM-like_dom
Gene 3D
ProteinModelPortal
H9J1W0
I0IYT1
A0A2A4JX89
A0A2H1VTG5
A0A212EHK6
A0A194PQ93
+ More
A0A0L7LH67 A0A084WB96 W5JI80 A0A182IT02 A0A2M4AFX5 A0A182V1J7 A0A182LPD5 A0A182HNH1 Q7Q355 A0A2M4BIQ3 A0A2P8ZLX4 A0A182WF88 A0A2M4BIS1 A0A182Q0R5 B0WBU6 A0A182T887 A0A182U5J1 A0A182YJZ9 A0A182PVD0 U5EYC5 A0A182MP96 A0A1L8DE05 A0A182FU18 A0A1B0CW50 E2A529 A0A310SVZ3 I0IYT2 A0A1B6FLL6 A0A1B6F907 D6W6W0 A0A1L8DEZ9 A0A088AUM5 A0A195CQH4 A0A1B6HR18 A0A1W4XGQ0 A0A1Y1LPH1 A0A151X3T6 A0A1W4XFI0 A0A1W4XGF1 A0A151IYH3 A0A1B6LGX7 A0A2J7PGJ2 A0A1B6DK64 A0A1W4XGQ4 A0A1B6L6U8 A0A158NKW0 A0A067R5D6 A0A151JYR5 K7IS83 A0A0L7R7S5 E0VQM9 A0A232ERE3 F4WE57 A0A088AJC7 A0A0L7RBV1 A0A1Y1LD93 E2AJ53 D6W847 A0A139WPG3 A0A0C9QWP9 A0A0P4WDL5 A0A151JVP9 A0A195BKU0 A0A158P455 K7IZF0 A0A195CNW3 A0A151J9B2 A0A2H1VA33 A0A151I5F2 A0A1W0WRE6 A0A1W4XMR4 A0A1W4XDI7 A0A1J1ICW3 A0A087V0H2 E9FVJ5 A0A2A4JTR3 E2BGG8 U4U133 N6TV38 T1JDD2 A0A1B6E3G3 A0A1B6DY20 A0A2W1CHU9 A0A151X0H6 E0VGF6 A0A0P5A7W7 A0A0P5JVG7 A0A0P5AWT0 A0A0P5VXK5 A0A0P5N487 A0A0P4Z4M9 A0A0P5NTE2 A0A0N8A344
A0A0L7LH67 A0A084WB96 W5JI80 A0A182IT02 A0A2M4AFX5 A0A182V1J7 A0A182LPD5 A0A182HNH1 Q7Q355 A0A2M4BIQ3 A0A2P8ZLX4 A0A182WF88 A0A2M4BIS1 A0A182Q0R5 B0WBU6 A0A182T887 A0A182U5J1 A0A182YJZ9 A0A182PVD0 U5EYC5 A0A182MP96 A0A1L8DE05 A0A182FU18 A0A1B0CW50 E2A529 A0A310SVZ3 I0IYT2 A0A1B6FLL6 A0A1B6F907 D6W6W0 A0A1L8DEZ9 A0A088AUM5 A0A195CQH4 A0A1B6HR18 A0A1W4XGQ0 A0A1Y1LPH1 A0A151X3T6 A0A1W4XFI0 A0A1W4XGF1 A0A151IYH3 A0A1B6LGX7 A0A2J7PGJ2 A0A1B6DK64 A0A1W4XGQ4 A0A1B6L6U8 A0A158NKW0 A0A067R5D6 A0A151JYR5 K7IS83 A0A0L7R7S5 E0VQM9 A0A232ERE3 F4WE57 A0A088AJC7 A0A0L7RBV1 A0A1Y1LD93 E2AJ53 D6W847 A0A139WPG3 A0A0C9QWP9 A0A0P4WDL5 A0A151JVP9 A0A195BKU0 A0A158P455 K7IZF0 A0A195CNW3 A0A151J9B2 A0A2H1VA33 A0A151I5F2 A0A1W0WRE6 A0A1W4XMR4 A0A1W4XDI7 A0A1J1ICW3 A0A087V0H2 E9FVJ5 A0A2A4JTR3 E2BGG8 U4U133 N6TV38 T1JDD2 A0A1B6E3G3 A0A1B6DY20 A0A2W1CHU9 A0A151X0H6 E0VGF6 A0A0P5A7W7 A0A0P5JVG7 A0A0P5AWT0 A0A0P5VXK5 A0A0P5N487 A0A0P4Z4M9 A0A0P5NTE2 A0A0N8A344
PDB
4M64
E-value=0.000583851,
Score=103
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
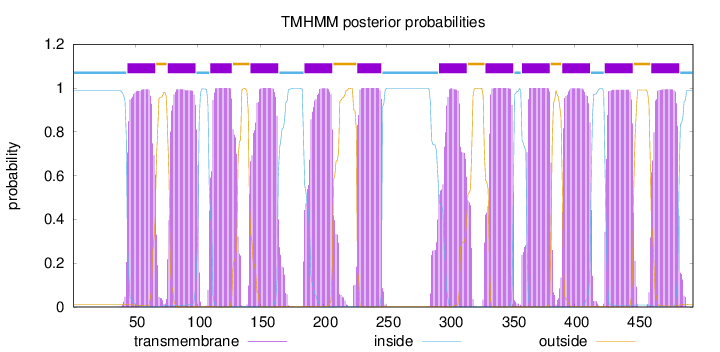
Length:
494
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
261.94816
Exp number, first 60 AAs:
16.55157
Total prob of N-in:
0.98766
POSSIBLE N-term signal
sequence
inside
1 - 43
TMhelix
44 - 66
outside
67 - 75
TMhelix
76 - 98
inside
99 - 109
TMhelix
110 - 127
outside
128 - 141
TMhelix
142 - 164
inside
165 - 184
TMhelix
185 - 207
outside
208 - 226
TMhelix
227 - 246
inside
247 - 291
TMhelix
292 - 314
outside
315 - 328
TMhelix
329 - 351
inside
352 - 357
TMhelix
358 - 380
outside
381 - 389
TMhelix
390 - 412
inside
413 - 423
TMhelix
424 - 446
outside
447 - 460
TMhelix
461 - 483
inside
484 - 494
Population Genetic Test Statistics
Pi
306.401542
Theta
161.550213
Tajima's D
0.325558
CLR
124.044858
CSRT
0.462776861156942
Interpretation
Uncertain
