Gene
KWMTBOMO02783
Pre Gene Modal
BGIBMGA003496
Annotation
PREDICTED:_GPI_ethanolamine_phosphate_transferase_1_[Amyelois_transitella]
Full name
GPI ethanolamine phosphate transferase 1
Alternative Name
MCD4 homolog
Phosphatidylinositol-glycan biosynthesis class N protein
Phosphatidylinositol-glycan biosynthesis class N protein
Location in the cell
PlasmaMembrane Reliability : 4.984
Sequence
CDS
ATGCCATTCTTAAGGTCTGTAGCAAATTCTAAAGGAAGATGGGGTATATCTAGAACTCAAGTTCCTACAGAGTCTAGGCCCGGTCATGTGGCTATTGCAGCCGGCTTTTATGAAGATCCTTCAGCTGTTGTAAAGGGGTGGAAAGAAAACCCAGTTGATTTTGATTCTGTATTCAACCAATCACAGTTCACTTGGTGTTGGGGTGCCTATGATATCATTAATATATTTACCAAAGACCATAATGCTGAAGAGGATGTAAATCTTCTTGCTAAATTGAAGAAAAATAAAATAGTATTTTTCTTGCACTTACTAGGAACTGATACATCTGGTCACACACACAAACCTCAAACAAGGAACTTTTTAACAACACTTAGATTTGTTGATGAAGGTATCAAAATTATAGAAGGAGTGATAAATACATTTTATAATGATGATGGAAAAACTACATTTCTTATGACTTCAGATCATGGAATGACAGACTGGGGATCACATGGTGCAGGCGATAATTATGAAACGGAGACGCCCTATGTGTTGTGGGGCTCTGGTGTGAACCAAGTTAATGAAACTAGACCGGACTCTGAAACTCTGCTTATGTCTTCAGAGCATAGATTTGATATTAACCAAATAGATTTGGCACCAATAATGGCAACGTTTATCTCTATACCGGTTCCTGTCAATTCTTTGGGTCAAGTGAAGACTGATTTATTGAACATGACACTGGCTAATAAAGCTAAAGCAGTTTATGGTAATAGTAGACAAATAGCTGCACAATACGATAAGAAGAGACATGATGTTGAATCAAATGCCATATCAATGCTTTACAAACCTTATGAACCACTGACCAAGAAAAAATACGAGGAAATAATAAATTTTGCAGAAACATTGTTAATTAAAGGAGAATATGAAAAGTTAATTATATGCAGTGAAGAAATCATGCAATTGAGTTTAAGAGGATTGAGATACTATAATGATTATTATCAAAAACCACTTCTTATTCTAGTTACTTTATCATTTATGGGATGGATTGTTTATTTATTGAAATTATTATCTGAACAAAAGATTCATACACATGCAGATACGTGTACAGCTAGTTGTGTAAGGGGAACTAAAATTCTAGTCAAGCAGTCTATTAATATTATTTTCTTAATTATTTTTGGATTTTCCATATACATAGTTTATGTGCAAAGTTTGCCAGTACAATATTTTGTATATTTTCTGATGCCCACTGCCTTGTGGTGGTATGCGATGTTATCAATGGAAGTTTGGTTTCAAATATACAAAAAAATTAAAAGGGACAAAGAACTGACATCAGTGCTATTACAAATTGTTTGTTATGGATTAGGGAGCATAGCAATGGGTTTATCATTTACCTACCGCTGGATGTTGAGCTTACCATTACTAGGTATGGCTCTGTGGCCATTCATGTCATCAACAAAATATCATTTAACTAATCCAGTGTTGTTGATCTGGTCAGGTGGATCTCTGCTACTTAGCATTTTTTCGTTCATGCCTGTTGTGGGAAAAAATGTGGTTATTTCATTAGTTATTGCTGCCGGCGTGCTTTGGATATTCGTGATAACAATATTCATTTACAAGATACTGATGCCGTTCCGTTACAATAAGGTTGAGAAACAAAGGGAAATGTATTTGTTCTCGATACAAATATTGTTATTACTTATTTCGTGTCATAACATTTACGTGCAGTCTAAACGGTTTGAACAAGGCACACCCGTATCAGCTTTCTATCAGACGGAAGCTTGGATTGTAGCAGTGACCTCATTGATCATGCCTTTCCTGTTTTCTAGAAGATTAATGTGTCGATTGATGGCGATAAATACGGGACTTATAAATTTCTATCTTTTGATGTCAGTTTCGCATGAAGGTCTGTTCATGGTTGCACTCATTATAAACGTAATGAGCTGGGTTTTTATTGAATTTAGACTTCTTCACGTGAAGGACGTAAAGATAATTGACTGCGCGTTTATTGAAGACACAAATGAAGAAAAAATTGGTCTTCAGATCGAAAGGAGTATTACTAGTCAAGACTTTAGAAGAGCTTTCTTTTTCGTATCCTTTACAAAATTGCGTCTAAAATATTATCATCAAATAATTCGACAAAGGTCCTCCATGTCTAAATCCACTGTTTTCAAGACACAAGTTGCGACCCTTAGAAATTACGCTGAACAACAAGAGCCGTACAGTTTGTCACTAAGGCACCACTTGGCTGTTTGA
Protein
MPFLRSVANSKGRWGISRTQVPTESRPGHVAIAAGFYEDPSAVVKGWKENPVDFDSVFNQSQFTWCWGAYDIINIFTKDHNAEEDVNLLAKLKKNKIVFFLHLLGTDTSGHTHKPQTRNFLTTLRFVDEGIKIIEGVINTFYNDDGKTTFLMTSDHGMTDWGSHGAGDNYETETPYVLWGSGVNQVNETRPDSETLLMSSEHRFDINQIDLAPIMATFISIPVPVNSLGQVKTDLLNMTLANKAKAVYGNSRQIAAQYDKKRHDVESNAISMLYKPYEPLTKKKYEEIINFAETLLIKGEYEKLIICSEEIMQLSLRGLRYYNDYYQKPLLILVTLSFMGWIVYLLKLLSEQKIHTHADTCTASCVRGTKILVKQSINIIFLIIFGFSIYIVYVQSLPVQYFVYFLMPTALWWYAMLSMEVWFQIYKKIKRDKELTSVLLQIVCYGLGSIAMGLSFTYRWMLSLPLLGMALWPFMSSTKYHLTNPVLLIWSGGSLLLSIFSFMPVVGKNVVISLVIAAGVLWIFVITIFIYKILMPFRYNKVEKQREMYLFSIQILLLLISCHNIYVQSKRFEQGTPVSAFYQTEAWIVAVTSLIMPFLFSRRLMCRLMAINTGLINFYLLMSVSHEGLFMVALIINVMSWVFIEFRLLHVKDVKIIDCAFIEDTNEEKIGLQIERSITSQDFRRAFFFVSFTKLRLKYYHQIIRQRSSMSKSTVFKTQVATLRNYAEQQEPYSLSLRHHLAV
Summary
Description
Ethanolamine phosphate transferase involved in glycosylphosphatidylinositol-anchor biosynthesis. Transfers ethanolamine phosphate to the first alpha-1,4-linked mannose of the glycosylphosphatidylinositol precursor of GPI-anchor (By similarity). May act as suppressor of replication stress and chromosome missegregation.
Similarity
Belongs to the PIGG/PIGN/PIGO family. PIGN subfamily.
Keywords
Complete proteome
Disease mutation
Endoplasmic reticulum
Epilepsy
Glycoprotein
GPI-anchor biosynthesis
Membrane
Polymorphism
Reference proteome
Transferase
Transmembrane
Transmembrane helix
Feature
chain GPI ethanolamine phosphate transferase 1
sequence variant In dbSNP:rs17069506.
sequence variant In dbSNP:rs17069506.
Uniprot
A0A2A4JXS1
A0A2H1VR59
A0A212ETA3
A0A194QVN5
A0A067RPH7
A0A2P8YAW0
+ More
A0A1B6FHP6 A0A210QQ19 A0A1B6L1H3 E7EXW2 A0A0L8HIE9 A0A1S3H469 A0A293LEF9 A0A2G8LAY8 V3ZGL6 R7U986 A0A147BRH6 A0A131XWR2 A0A250YLR4 A0A2K5RAR6 A0A2D0PTK1 A0A2J8NDD0 A0A2K6T6D5 H0WT04 A0A2K5RAV8 A0A2K5EY07 A0A2K6T6J1 A0A2K6T6I7 A0A2K6CJP7 A0A2K5EXV6 A0A1B6HFW5 A0A1E1X5F0 A0A0F7ZDT4 A0A2K5EXY4 A0A1W2PQZ1 G1PUC4 A0A131Z2N9 A0A2I3SK40 A0A2J8W3G6 A0A1E1XPK6 A0A224YVQ4 A0A1S2ZTE0 L7M6U1 A0A2I2ZJG6 H9EVK7 A0A2R8M9W8 A0A2K5VNQ4 A0A2K6MMP5 A0A131XAH2 A0A2I3TGD6 A0A0D9S006 G7PWW5 G3QQS9 A0A2I3TW05 A0A2K5J9X8 A0A2K6NNF1 A0A2K5YX69 A0A2R8PL21 A0A2R9BVL4 A0A2I3S836 S7NIZ4 A0A3N0Y917 A0A2J8NDD7 A0A2I2ZW78 A0A2K5YWP8 A0A2K6NND4 A0A2I3RAS4 A0A2J8NDE3 A0A2J8NDE5 A0A2K5VNI4 A0A2K6MMI4 A0A2K5NYQ2 A0A2K5JA28 A0A1W2PNR0 I3MD84 A0A2K5YWT2 A0A2K6NNE1 A0A2K5NYN8 A0A024R2C3 O95427 A0A2K6CKU1 G3SR92 B2RCI8 A0A1W2PS19 F7FX72 A0A2K5NYU2 A0A1B6C8V4 A0A2K6CKL3 A0A1B6CUA4 A0A2K5NYU5 A0A2J8W3E9 A0A2J8W3E0 A0A1W2PQA9 A0A2J8W3I5 A0A0P6J835 A0A2K6CKK8 G1RB70
A0A1B6FHP6 A0A210QQ19 A0A1B6L1H3 E7EXW2 A0A0L8HIE9 A0A1S3H469 A0A293LEF9 A0A2G8LAY8 V3ZGL6 R7U986 A0A147BRH6 A0A131XWR2 A0A250YLR4 A0A2K5RAR6 A0A2D0PTK1 A0A2J8NDD0 A0A2K6T6D5 H0WT04 A0A2K5RAV8 A0A2K5EY07 A0A2K6T6J1 A0A2K6T6I7 A0A2K6CJP7 A0A2K5EXV6 A0A1B6HFW5 A0A1E1X5F0 A0A0F7ZDT4 A0A2K5EXY4 A0A1W2PQZ1 G1PUC4 A0A131Z2N9 A0A2I3SK40 A0A2J8W3G6 A0A1E1XPK6 A0A224YVQ4 A0A1S2ZTE0 L7M6U1 A0A2I2ZJG6 H9EVK7 A0A2R8M9W8 A0A2K5VNQ4 A0A2K6MMP5 A0A131XAH2 A0A2I3TGD6 A0A0D9S006 G7PWW5 G3QQS9 A0A2I3TW05 A0A2K5J9X8 A0A2K6NNF1 A0A2K5YX69 A0A2R8PL21 A0A2R9BVL4 A0A2I3S836 S7NIZ4 A0A3N0Y917 A0A2J8NDD7 A0A2I2ZW78 A0A2K5YWP8 A0A2K6NND4 A0A2I3RAS4 A0A2J8NDE3 A0A2J8NDE5 A0A2K5VNI4 A0A2K6MMI4 A0A2K5NYQ2 A0A2K5JA28 A0A1W2PNR0 I3MD84 A0A2K5YWT2 A0A2K6NNE1 A0A2K5NYN8 A0A024R2C3 O95427 A0A2K6CKU1 G3SR92 B2RCI8 A0A1W2PS19 F7FX72 A0A2K5NYU2 A0A1B6C8V4 A0A2K6CKL3 A0A1B6CUA4 A0A2K5NYU5 A0A2J8W3E9 A0A2J8W3E0 A0A1W2PQA9 A0A2J8W3I5 A0A0P6J835 A0A2K6CKK8 G1RB70
EC Number
2.-.-.-
Pubmed
EMBL
NWSH01000438
PCG76434.1
ODYU01003919
SOQ43278.1
AGBW02012631
OWR44684.1
+ More
KQ461181 KPJ07616.1 KK852502 KDR22535.1 PYGN01000747 PSN41373.1 GECZ01020052 JAS49717.1 NEDP02002420 OWF50840.1 GEBQ01022439 JAT17538.1 BX640462 KQ418168 KOF88545.1 GFWV01001519 MAA26249.1 MRZV01000143 PIK57412.1 KB202444 ESO90338.1 AMQN01009923 KB306612 ELT99695.1 GEGO01002048 JAR93356.1 GEFM01004092 JAP71704.1 GFFW01000231 JAV44557.1 NBAG03000231 PNI69778.1 AAQR03056653 AAQR03056654 AAQR03056655 AAQR03056656 AAQR03056657 GECU01034126 JAS73580.1 GFAC01004740 JAT94448.1 GBEX01001431 JAI13129.1 AC090354 AC090396 AC105183 FP340563 KF456430 AAPE02014648 GEDV01003831 JAP84726.1 AACZ04062428 NDHI03003401 PNJ64302.1 GFAA01002187 JAU01248.1 GFPF01010551 MAA21697.1 GACK01005269 JAA59765.1 CABD030109837 CABD030109838 CABD030109839 CABD030109840 CABD030109841 CABD030109842 CABD030109843 CABD030109844 CABD030109845 JU322660 AFE66416.1 AQIA01031925 AQIA01031926 AQIA01031927 AQIA01031928 AQIA01031929 GEFH01004498 JAP64083.1 PNI69781.1 AQIB01127725 AQIB01127726 CM001293 EHH58938.1 AJFE02095763 AJFE02095764 AJFE02095765 AJFE02095766 AJFE02095767 GABC01005387 GABF01003366 JAA05951.1 JAA18779.1 PNI69755.1 PNI69757.1 PNI69760.1 PNI69766.1 PNI69767.1 PNI69768.1 PNI69788.1 KE164355 EPQ17389.1 RJVU01049572 ROL42647.1 PNI69790.1 PNI69784.1 PNI69773.1 PNI69785.1 AGTP01028278 AGTP01028279 CH471096 EAW63119.1 AF109219 BC028363 AL137607 AK315134 BAG37585.1 GEDC01027361 JAS09937.1 GEDC01020232 JAS17066.1 PNJ64305.1 PNJ64281.1 PNJ64283.1 PNJ64284.1 PNJ64285.1 PNJ64293.1 PNJ64294.1 PNJ64303.1 PNJ64306.1 GEBF01003895 JAN99737.1 ADFV01021533 ADFV01021534 ADFV01021535 ADFV01021536 ADFV01021537 ADFV01021538 ADFV01021539 ADFV01021540
KQ461181 KPJ07616.1 KK852502 KDR22535.1 PYGN01000747 PSN41373.1 GECZ01020052 JAS49717.1 NEDP02002420 OWF50840.1 GEBQ01022439 JAT17538.1 BX640462 KQ418168 KOF88545.1 GFWV01001519 MAA26249.1 MRZV01000143 PIK57412.1 KB202444 ESO90338.1 AMQN01009923 KB306612 ELT99695.1 GEGO01002048 JAR93356.1 GEFM01004092 JAP71704.1 GFFW01000231 JAV44557.1 NBAG03000231 PNI69778.1 AAQR03056653 AAQR03056654 AAQR03056655 AAQR03056656 AAQR03056657 GECU01034126 JAS73580.1 GFAC01004740 JAT94448.1 GBEX01001431 JAI13129.1 AC090354 AC090396 AC105183 FP340563 KF456430 AAPE02014648 GEDV01003831 JAP84726.1 AACZ04062428 NDHI03003401 PNJ64302.1 GFAA01002187 JAU01248.1 GFPF01010551 MAA21697.1 GACK01005269 JAA59765.1 CABD030109837 CABD030109838 CABD030109839 CABD030109840 CABD030109841 CABD030109842 CABD030109843 CABD030109844 CABD030109845 JU322660 AFE66416.1 AQIA01031925 AQIA01031926 AQIA01031927 AQIA01031928 AQIA01031929 GEFH01004498 JAP64083.1 PNI69781.1 AQIB01127725 AQIB01127726 CM001293 EHH58938.1 AJFE02095763 AJFE02095764 AJFE02095765 AJFE02095766 AJFE02095767 GABC01005387 GABF01003366 JAA05951.1 JAA18779.1 PNI69755.1 PNI69757.1 PNI69760.1 PNI69766.1 PNI69767.1 PNI69768.1 PNI69788.1 KE164355 EPQ17389.1 RJVU01049572 ROL42647.1 PNI69790.1 PNI69784.1 PNI69773.1 PNI69785.1 AGTP01028278 AGTP01028279 CH471096 EAW63119.1 AF109219 BC028363 AL137607 AK315134 BAG37585.1 GEDC01027361 JAS09937.1 GEDC01020232 JAS17066.1 PNJ64305.1 PNJ64281.1 PNJ64283.1 PNJ64284.1 PNJ64285.1 PNJ64293.1 PNJ64294.1 PNJ64303.1 PNJ64306.1 GEBF01003895 JAN99737.1 ADFV01021533 ADFV01021534 ADFV01021535 ADFV01021536 ADFV01021537 ADFV01021538 ADFV01021539 ADFV01021540
Proteomes
UP000218220
UP000007151
UP000053240
UP000027135
UP000245037
UP000242188
+ More
UP000000437 UP000053454 UP000085678 UP000230750 UP000030746 UP000014760 UP000233040 UP000221080 UP000233220 UP000005225 UP000233020 UP000233120 UP000005640 UP000001074 UP000002277 UP000079721 UP000001519 UP000008225 UP000233100 UP000233180 UP000029965 UP000009130 UP000233080 UP000233200 UP000233140 UP000240080 UP000233060 UP000005215 UP000007646 UP000002280 UP000001073
UP000000437 UP000053454 UP000085678 UP000230750 UP000030746 UP000014760 UP000233040 UP000221080 UP000233220 UP000005225 UP000233020 UP000233120 UP000005640 UP000001074 UP000002277 UP000079721 UP000001519 UP000008225 UP000233100 UP000233180 UP000029965 UP000009130 UP000233080 UP000233200 UP000233140 UP000240080 UP000233060 UP000005215 UP000007646 UP000002280 UP000001073
Interpro
SUPFAM
SSF53649
SSF53649
Gene 3D
ProteinModelPortal
A0A2A4JXS1
A0A2H1VR59
A0A212ETA3
A0A194QVN5
A0A067RPH7
A0A2P8YAW0
+ More
A0A1B6FHP6 A0A210QQ19 A0A1B6L1H3 E7EXW2 A0A0L8HIE9 A0A1S3H469 A0A293LEF9 A0A2G8LAY8 V3ZGL6 R7U986 A0A147BRH6 A0A131XWR2 A0A250YLR4 A0A2K5RAR6 A0A2D0PTK1 A0A2J8NDD0 A0A2K6T6D5 H0WT04 A0A2K5RAV8 A0A2K5EY07 A0A2K6T6J1 A0A2K6T6I7 A0A2K6CJP7 A0A2K5EXV6 A0A1B6HFW5 A0A1E1X5F0 A0A0F7ZDT4 A0A2K5EXY4 A0A1W2PQZ1 G1PUC4 A0A131Z2N9 A0A2I3SK40 A0A2J8W3G6 A0A1E1XPK6 A0A224YVQ4 A0A1S2ZTE0 L7M6U1 A0A2I2ZJG6 H9EVK7 A0A2R8M9W8 A0A2K5VNQ4 A0A2K6MMP5 A0A131XAH2 A0A2I3TGD6 A0A0D9S006 G7PWW5 G3QQS9 A0A2I3TW05 A0A2K5J9X8 A0A2K6NNF1 A0A2K5YX69 A0A2R8PL21 A0A2R9BVL4 A0A2I3S836 S7NIZ4 A0A3N0Y917 A0A2J8NDD7 A0A2I2ZW78 A0A2K5YWP8 A0A2K6NND4 A0A2I3RAS4 A0A2J8NDE3 A0A2J8NDE5 A0A2K5VNI4 A0A2K6MMI4 A0A2K5NYQ2 A0A2K5JA28 A0A1W2PNR0 I3MD84 A0A2K5YWT2 A0A2K6NNE1 A0A2K5NYN8 A0A024R2C3 O95427 A0A2K6CKU1 G3SR92 B2RCI8 A0A1W2PS19 F7FX72 A0A2K5NYU2 A0A1B6C8V4 A0A2K6CKL3 A0A1B6CUA4 A0A2K5NYU5 A0A2J8W3E9 A0A2J8W3E0 A0A1W2PQA9 A0A2J8W3I5 A0A0P6J835 A0A2K6CKK8 G1RB70
A0A1B6FHP6 A0A210QQ19 A0A1B6L1H3 E7EXW2 A0A0L8HIE9 A0A1S3H469 A0A293LEF9 A0A2G8LAY8 V3ZGL6 R7U986 A0A147BRH6 A0A131XWR2 A0A250YLR4 A0A2K5RAR6 A0A2D0PTK1 A0A2J8NDD0 A0A2K6T6D5 H0WT04 A0A2K5RAV8 A0A2K5EY07 A0A2K6T6J1 A0A2K6T6I7 A0A2K6CJP7 A0A2K5EXV6 A0A1B6HFW5 A0A1E1X5F0 A0A0F7ZDT4 A0A2K5EXY4 A0A1W2PQZ1 G1PUC4 A0A131Z2N9 A0A2I3SK40 A0A2J8W3G6 A0A1E1XPK6 A0A224YVQ4 A0A1S2ZTE0 L7M6U1 A0A2I2ZJG6 H9EVK7 A0A2R8M9W8 A0A2K5VNQ4 A0A2K6MMP5 A0A131XAH2 A0A2I3TGD6 A0A0D9S006 G7PWW5 G3QQS9 A0A2I3TW05 A0A2K5J9X8 A0A2K6NNF1 A0A2K5YX69 A0A2R8PL21 A0A2R9BVL4 A0A2I3S836 S7NIZ4 A0A3N0Y917 A0A2J8NDD7 A0A2I2ZW78 A0A2K5YWP8 A0A2K6NND4 A0A2I3RAS4 A0A2J8NDE3 A0A2J8NDE5 A0A2K5VNI4 A0A2K6MMI4 A0A2K5NYQ2 A0A2K5JA28 A0A1W2PNR0 I3MD84 A0A2K5YWT2 A0A2K6NNE1 A0A2K5NYN8 A0A024R2C3 O95427 A0A2K6CKU1 G3SR92 B2RCI8 A0A1W2PS19 F7FX72 A0A2K5NYU2 A0A1B6C8V4 A0A2K6CKL3 A0A1B6CUA4 A0A2K5NYU5 A0A2J8W3E9 A0A2J8W3E0 A0A1W2PQA9 A0A2J8W3I5 A0A0P6J835 A0A2K6CKK8 G1RB70
Ontologies
PATHWAY
GO
PANTHER
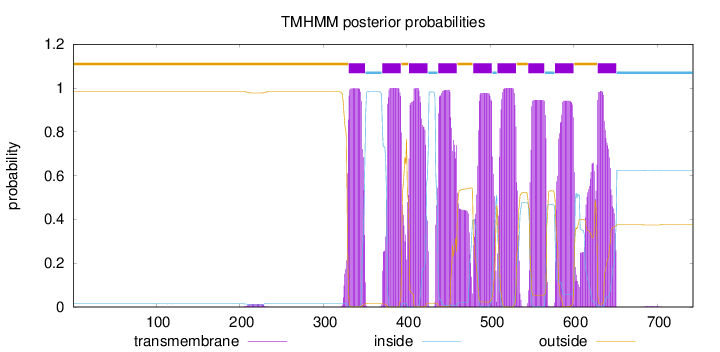
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
743
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
202.16691
Exp number, first 60 AAs:
3e-05
Total prob of N-in:
0.01452
outside
1 - 330
TMhelix
331 - 350
inside
351 - 370
TMhelix
371 - 393
outside
394 - 402
TMhelix
403 - 425
inside
426 - 437
TMhelix
438 - 460
outside
461 - 479
TMhelix
480 - 502
inside
503 - 508
TMhelix
509 - 531
outside
532 - 545
TMhelix
546 - 565
inside
566 - 577
TMhelix
578 - 600
outside
601 - 628
TMhelix
629 - 651
inside
652 - 743
Population Genetic Test Statistics
Pi
265.070454
Theta
19.597392
Tajima's D
0.454418
CLR
1.919906
CSRT
0.508924553772311
Interpretation
Uncertain
