Gene
KWMTBOMO02780
Pre Gene Modal
BGIBMGA003701
Annotation
PREDICTED:_metallophosphoesterase_1_homolog_isoform_X2_[Bombyx_mori]
Full name
Metallophosphoesterase 1
+ More
Metallophosphoesterase 1 homolog
Metallophosphoesterase 1 homolog
Alternative Name
Post-GPI attachment to proteins factor 5
Post-GPI attachment to proteins 5 ortholog
Post-GPI attachment to proteins 5 ortholog
Location in the cell
Nuclear Reliability : 3.245
Sequence
CDS
ATGTTGCAGTCATTAAAAAGTGCGGTCGAATATGTATCTACAGAAGATTTGAAGAAGTGGAAGAAGGTGGTAACGAAACGCAGAAAAGTAATGAAAGAAGATTGGGAGCGGATCGTCCGATTTGATAATATTTGCGATCAAGAGCTTATAATAAATTTACAAGATTGTTTTAGCGATGCAGAAACTGATTCAGAAATATATTTGTGGTTTATAGCGCAGTGTAATTGGCCTGTTACAAATGACACACATAAAAGTATACAGAGTTTGAAAGCCCTTATGATATCAGACACCCATCTACTGGGACCAAGAAGAGGGCATTGGCTGGATAAAATGAGACGCGAGTGGCAGATGCATCAAGCATTTCAAACTATAATGATGATGCTGAGCCCTGACGTTGTATTTGTTTTAGGTGATTTATTTGATGAAGGAGAATGGACAAATAATAAAGAGTTTCAAGAGTATGTTGAACGTTTCTATAGATTATTTATGGTACCACCCCACGTGAAGATGTATGTTGTTGCAGGGAATCATGATATTGGATTTCATAATTACATAAGAAAGGGAGCTATCCAAAGATTTTACAAGCTCCTGAACACGTCATCAGTTGAATTTGTCACTTTGAAAGACATTCATTTTGTACTTTTGAATTCAATGGCAATGGAGGGAGATTCCTGTCAATTGTGTAGTGAAGCACGGAATGATATACAACATATATCAGGAAGACTAAAATGTTCAGAGGACCCTAAGTATTGTTTCGGAAATAAATTCAAATCCCTGAATTATAGTCGTCCAATCCTTTTACAGCATTTCCCATTATACAGAAAATCTGATTCAATGTGTACAGAACCAGATTCACCGCCATTGCCTGAACGGGACAAGCCGTTCAGACTCAAAATCGACGCTCTGTCTAAAGAGGCTTCAGAGTTCCTAGTAAGCAAAATCAAACCAAGGGCTGTTTTCGGAGGGCACACACATCACGGTTGTCTTTTACATCACAAATACGAAAATCTAGGGCAGCTAGAATTTAAGGAGTATTCAATACCATCATTCTCTTGGAGAAATAGACCAGATCCAAAGTATATGTTGGTAACATTAACGCCAAACGACTACAACGTACAGAAATGTGGACTACCTCAAGAAGCTACGATAGTCATTACTGCTATATCAATGCTTCTAATACTTGCATGTTTTATTGCAAAGAGGAAATATATAGGAAGGTGA
Protein
MLQSLKSAVEYVSTEDLKKWKKVVTKRRKVMKEDWERIVRFDNICDQELIINLQDCFSDAETDSEIYLWFIAQCNWPVTNDTHKSIQSLKALMISDTHLLGPRRGHWLDKMRREWQMHQAFQTIMMMLSPDVVFVLGDLFDEGEWTNNKEFQEYVERFYRLFMVPPHVKMYVVAGNHDIGFHNYIRKGAIQRFYKLLNTSSVEFVTLKDIHFVLLNSMAMEGDSCQLCSEARNDIQHISGRLKCSEDPKYCFGNKFKSLNYSRPILLQHFPLYRKSDSMCTEPDSPPLPERDKPFRLKIDALSKEASEFLVSKIKPRAVFGGHTHHGCLLHHKYENLGQLEFKEYSIPSFSWRNRPDPKYMLVTLTPNDYNVQKCGLPQEATIVITAISMLLILACFIAKRKYIGR
Summary
Description
Metallophosphoesterase required for transport of GPI-anchor proteins from the endoplasmic reticulum to the Golgi. Acts in lipid remodeling steps of GPI-anchor maturation by mediating the removal of a side-chain ethanolamine-phosphate (EtNP) from the second Man (Man2) of the GPI intermediate, an essential step for efficient transport of GPI-anchor proteins (By similarity).
Metallophosphoesterase.
Metallophosphoesterase.
Cofactor
Mn(2+)
Subunit
Interacts with GPI-anchor proteins. Interacts with TMED10 (By similarity).
Similarity
Belongs to the metallophosphoesterase superfamily. MPPE1 family.
Keywords
Complete proteome
ER-Golgi transport
Golgi apparatus
GPI-anchor biosynthesis
Hydrolase
Manganese
Membrane
Metal-binding
Reference proteome
Transmembrane
Transmembrane helix
Transport
Alternative splicing
Feature
chain Metallophosphoesterase 1
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J2G4
A0A2H1W5L6
A0A212ET80
A0A194PN91
A0A2A4JFF8
A0A2H1X198
+ More
A0A0A9WNJ7 A0A0A9WS93 A0A023F336 A0A194PU72 A0A0N0PC24 V5HTK6 J9L1W7 A0A1B6C6R9 E0VC39 A0A2J7Q7Q9 A0A067RJK9 A0A3Q0IKV5 A0A224YYP0 L7M1A4 F6XU00 A0A310SK47 A0A088A1E3 V5HDR6 A0A2I2UL78 A0A1S4E770 A0A131YFK5 A0A0L7QVY0 A0A182Q9E5 A0A1D2NFJ1 A0A1Y1KCC5 E2C2S4 B4MVX2 A0A2Y9GHU4 A0A2U3WKS5 A0A2U3YZD9 A0A0M9A4P4 A0A2A3EIH6 R0LK38 A0A2P6LB90 M3XZJ6 U3IYE3 A0A224XFU0 A0A0P5MQY5 Q16HW6 A0A0P6DQT1 A0A182VUJ8 A0A1S4G0A0 A0A0P6HB29 A0A0P6FGV9 A0A0N8DJW2 G3MSF5 A0A023GDZ4 A0A0P5ZSE6 A0A182VAK9 Q7PVE7 A0A182T103 A0A1S4H1I0 A0A1Z5LAN5 G1LNH7 A0A226F3Q2 D2I2M6 W5J8Y5 B3N716 A0A0Q5WBM3 A0A182LY21 A0A182MZ21 F1LBC3 A0A232F6G9 T1EB03 A0A093LAM1 A0A1Z5L2V3 A0A0K8V4N5 A0A094LG59 A0A2Y9IRJ0 L5LW21 A0A212F733 A0A182P9G5 B4Q6B2 A0A091MPY0 A0A2M4BRP8 A0A0J9QYE7 B3MPI5 F1KS81 A0A0K8TT47 A0A2M4BRB8 Q9VLR9 Q9VLR9-2 K7IQV4 A0A034WAC4 S7MXE3 A0A2R2MIX8 A0A0P6J2Q9 A0A099ZM14 F4WK18 A0A3L8DLL8 A0A0B6ZV66 G1PHL5 A0A2Y9IVG2
A0A0A9WNJ7 A0A0A9WS93 A0A023F336 A0A194PU72 A0A0N0PC24 V5HTK6 J9L1W7 A0A1B6C6R9 E0VC39 A0A2J7Q7Q9 A0A067RJK9 A0A3Q0IKV5 A0A224YYP0 L7M1A4 F6XU00 A0A310SK47 A0A088A1E3 V5HDR6 A0A2I2UL78 A0A1S4E770 A0A131YFK5 A0A0L7QVY0 A0A182Q9E5 A0A1D2NFJ1 A0A1Y1KCC5 E2C2S4 B4MVX2 A0A2Y9GHU4 A0A2U3WKS5 A0A2U3YZD9 A0A0M9A4P4 A0A2A3EIH6 R0LK38 A0A2P6LB90 M3XZJ6 U3IYE3 A0A224XFU0 A0A0P5MQY5 Q16HW6 A0A0P6DQT1 A0A182VUJ8 A0A1S4G0A0 A0A0P6HB29 A0A0P6FGV9 A0A0N8DJW2 G3MSF5 A0A023GDZ4 A0A0P5ZSE6 A0A182VAK9 Q7PVE7 A0A182T103 A0A1S4H1I0 A0A1Z5LAN5 G1LNH7 A0A226F3Q2 D2I2M6 W5J8Y5 B3N716 A0A0Q5WBM3 A0A182LY21 A0A182MZ21 F1LBC3 A0A232F6G9 T1EB03 A0A093LAM1 A0A1Z5L2V3 A0A0K8V4N5 A0A094LG59 A0A2Y9IRJ0 L5LW21 A0A212F733 A0A182P9G5 B4Q6B2 A0A091MPY0 A0A2M4BRP8 A0A0J9QYE7 B3MPI5 F1KS81 A0A0K8TT47 A0A2M4BRB8 Q9VLR9 Q9VLR9-2 K7IQV4 A0A034WAC4 S7MXE3 A0A2R2MIX8 A0A0P6J2Q9 A0A099ZM14 F4WK18 A0A3L8DLL8 A0A0B6ZV66 G1PHL5 A0A2Y9IVG2
EC Number
3.1.-.-
Pubmed
19121390
22118469
26354079
25401762
26823975
25474469
+ More
25765539 20566863 24845553 28797301 25576852 16341006 17975172 26830274 27289101 28004739 20798317 17994087 23749191 17510324 22216098 12364791 28528879 20010809 20920257 23761445 18057021 21685128 28648823 22936249 26369729 10731132 12537572 12537569 20075255 25348373 21719571 30249741 21993624
25765539 20566863 24845553 28797301 25576852 16341006 17975172 26830274 27289101 28004739 20798317 17994087 23749191 17510324 22216098 12364791 28528879 20010809 20920257 23761445 18057021 21685128 28648823 22936249 26369729 10731132 12537572 12537569 20075255 25348373 21719571 30249741 21993624
EMBL
BABH01007923
ODYU01006495
SOQ48380.1
AGBW02012631
OWR44687.1
KQ459597
+ More
KPI94901.1 NWSH01001661 PCG70506.1 ODYU01012677 SOQ59115.1 GBHO01034250 GDHC01015120 JAG09354.1 JAQ03509.1 GBHO01034251 GDHC01003712 JAG09353.1 JAQ14917.1 GBBI01003176 JAC15536.1 KQ459598 KPI94685.1 KQ460709 KPJ12723.1 GANP01002734 JAB81734.1 ABLF02014628 GEDC01028131 JAS09167.1 DS235047 EEB10945.1 NEVH01017440 PNF24623.1 KK852432 KDR24002.1 GFPF01009673 MAA20819.1 GACK01007402 JAA57632.1 AAEX03005517 KQ765234 OAD54015.1 GANP01002733 JAB81735.1 AANG04003715 GEDV01010518 JAP78039.1 KQ414722 KOC62711.1 AXCN02000792 LJIJ01000061 ODN03855.1 GEZM01091105 JAV57096.1 GL452203 EFN77759.1 CH963857 EDW75842.1 KQ435748 KOX76406.1 KZ288250 PBC31082.1 KB742642 EOB06064.1 MWRG01000528 PRD35857.1 AEYP01011404 AEYP01011405 AEYP01011406 ADON01029504 GFTR01005089 JAW11337.1 GDIQ01152632 JAK99093.1 CH478133 EAT33852.1 GDIQ01074058 JAN20679.1 GDIQ01031418 JAN63319.1 GDIQ01057462 JAN37275.1 GDIP01026551 JAM77164.1 JO844806 AEO36423.1 GBBM01003346 JAC32072.1 GDIP01039974 LRGB01000084 JAM63741.1 KZS21114.1 AAAB01008984 EAA14828.3 GFJQ02002722 JAW04248.1 ACTA01116556 LNIX01000001 OXA63821.1 GL194190 ADMH02002094 ETN59264.1 CH954177 EDV59312.1 KQS70748.1 KQS70749.1 AXCM01002487 JI176273 ADY47427.1 NNAY01000802 OXU26431.1 GAMD01000548 JAB01043.1 KK569097 KFW06256.1 GFJQ02005576 JAW01394.1 GDHF01018749 JAI33565.1 KL357111 KFZ62954.1 KB106856 ELK30624.1 AGBW02009947 OWR49519.1 CM000361 CM002910 EDX04188.1 KMY88971.1 KK834900 KFP79291.1 GGFJ01006598 MBW55739.1 KMY88973.1 CH902620 EDV31281.1 JI165018 ADY40735.1 GDAI01000280 JAI17323.1 GGFJ01006469 MBW55610.1 AE014134 AY094655 GAKP01006416 JAC52536.1 KE162428 EPQ08200.1 GEBF01006062 JAN97570.1 KL894559 KGL81850.1 GL888190 EGI65468.1 QOIP01000007 RLU21082.1 HACG01025634 CEK72499.1 AAPE02018921
KPI94901.1 NWSH01001661 PCG70506.1 ODYU01012677 SOQ59115.1 GBHO01034250 GDHC01015120 JAG09354.1 JAQ03509.1 GBHO01034251 GDHC01003712 JAG09353.1 JAQ14917.1 GBBI01003176 JAC15536.1 KQ459598 KPI94685.1 KQ460709 KPJ12723.1 GANP01002734 JAB81734.1 ABLF02014628 GEDC01028131 JAS09167.1 DS235047 EEB10945.1 NEVH01017440 PNF24623.1 KK852432 KDR24002.1 GFPF01009673 MAA20819.1 GACK01007402 JAA57632.1 AAEX03005517 KQ765234 OAD54015.1 GANP01002733 JAB81735.1 AANG04003715 GEDV01010518 JAP78039.1 KQ414722 KOC62711.1 AXCN02000792 LJIJ01000061 ODN03855.1 GEZM01091105 JAV57096.1 GL452203 EFN77759.1 CH963857 EDW75842.1 KQ435748 KOX76406.1 KZ288250 PBC31082.1 KB742642 EOB06064.1 MWRG01000528 PRD35857.1 AEYP01011404 AEYP01011405 AEYP01011406 ADON01029504 GFTR01005089 JAW11337.1 GDIQ01152632 JAK99093.1 CH478133 EAT33852.1 GDIQ01074058 JAN20679.1 GDIQ01031418 JAN63319.1 GDIQ01057462 JAN37275.1 GDIP01026551 JAM77164.1 JO844806 AEO36423.1 GBBM01003346 JAC32072.1 GDIP01039974 LRGB01000084 JAM63741.1 KZS21114.1 AAAB01008984 EAA14828.3 GFJQ02002722 JAW04248.1 ACTA01116556 LNIX01000001 OXA63821.1 GL194190 ADMH02002094 ETN59264.1 CH954177 EDV59312.1 KQS70748.1 KQS70749.1 AXCM01002487 JI176273 ADY47427.1 NNAY01000802 OXU26431.1 GAMD01000548 JAB01043.1 KK569097 KFW06256.1 GFJQ02005576 JAW01394.1 GDHF01018749 JAI33565.1 KL357111 KFZ62954.1 KB106856 ELK30624.1 AGBW02009947 OWR49519.1 CM000361 CM002910 EDX04188.1 KMY88971.1 KK834900 KFP79291.1 GGFJ01006598 MBW55739.1 KMY88973.1 CH902620 EDV31281.1 JI165018 ADY40735.1 GDAI01000280 JAI17323.1 GGFJ01006469 MBW55610.1 AE014134 AY094655 GAKP01006416 JAC52536.1 KE162428 EPQ08200.1 GEBF01006062 JAN97570.1 KL894559 KGL81850.1 GL888190 EGI65468.1 QOIP01000007 RLU21082.1 HACG01025634 CEK72499.1 AAPE02018921
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000053240
UP000007819
+ More
UP000009046 UP000235965 UP000027135 UP000079169 UP000002254 UP000005203 UP000011712 UP000053825 UP000075886 UP000094527 UP000008237 UP000007798 UP000248481 UP000245340 UP000245341 UP000053105 UP000242457 UP000000715 UP000016666 UP000008820 UP000075920 UP000076858 UP000075903 UP000007062 UP000075901 UP000008912 UP000198287 UP000000673 UP000008711 UP000075883 UP000075884 UP000215335 UP000248482 UP000075885 UP000000304 UP000007801 UP000000803 UP000002358 UP000085678 UP000053641 UP000007755 UP000279307 UP000001074
UP000009046 UP000235965 UP000027135 UP000079169 UP000002254 UP000005203 UP000011712 UP000053825 UP000075886 UP000094527 UP000008237 UP000007798 UP000248481 UP000245340 UP000245341 UP000053105 UP000242457 UP000000715 UP000016666 UP000008820 UP000075920 UP000076858 UP000075903 UP000007062 UP000075901 UP000008912 UP000198287 UP000000673 UP000008711 UP000075883 UP000075884 UP000215335 UP000248482 UP000075885 UP000000304 UP000007801 UP000000803 UP000002358 UP000085678 UP000053641 UP000007755 UP000279307 UP000001074
Interpro
SUPFAM
SSF100950
SSF100950
Gene 3D
ProteinModelPortal
H9J2G4
A0A2H1W5L6
A0A212ET80
A0A194PN91
A0A2A4JFF8
A0A2H1X198
+ More
A0A0A9WNJ7 A0A0A9WS93 A0A023F336 A0A194PU72 A0A0N0PC24 V5HTK6 J9L1W7 A0A1B6C6R9 E0VC39 A0A2J7Q7Q9 A0A067RJK9 A0A3Q0IKV5 A0A224YYP0 L7M1A4 F6XU00 A0A310SK47 A0A088A1E3 V5HDR6 A0A2I2UL78 A0A1S4E770 A0A131YFK5 A0A0L7QVY0 A0A182Q9E5 A0A1D2NFJ1 A0A1Y1KCC5 E2C2S4 B4MVX2 A0A2Y9GHU4 A0A2U3WKS5 A0A2U3YZD9 A0A0M9A4P4 A0A2A3EIH6 R0LK38 A0A2P6LB90 M3XZJ6 U3IYE3 A0A224XFU0 A0A0P5MQY5 Q16HW6 A0A0P6DQT1 A0A182VUJ8 A0A1S4G0A0 A0A0P6HB29 A0A0P6FGV9 A0A0N8DJW2 G3MSF5 A0A023GDZ4 A0A0P5ZSE6 A0A182VAK9 Q7PVE7 A0A182T103 A0A1S4H1I0 A0A1Z5LAN5 G1LNH7 A0A226F3Q2 D2I2M6 W5J8Y5 B3N716 A0A0Q5WBM3 A0A182LY21 A0A182MZ21 F1LBC3 A0A232F6G9 T1EB03 A0A093LAM1 A0A1Z5L2V3 A0A0K8V4N5 A0A094LG59 A0A2Y9IRJ0 L5LW21 A0A212F733 A0A182P9G5 B4Q6B2 A0A091MPY0 A0A2M4BRP8 A0A0J9QYE7 B3MPI5 F1KS81 A0A0K8TT47 A0A2M4BRB8 Q9VLR9 Q9VLR9-2 K7IQV4 A0A034WAC4 S7MXE3 A0A2R2MIX8 A0A0P6J2Q9 A0A099ZM14 F4WK18 A0A3L8DLL8 A0A0B6ZV66 G1PHL5 A0A2Y9IVG2
A0A0A9WNJ7 A0A0A9WS93 A0A023F336 A0A194PU72 A0A0N0PC24 V5HTK6 J9L1W7 A0A1B6C6R9 E0VC39 A0A2J7Q7Q9 A0A067RJK9 A0A3Q0IKV5 A0A224YYP0 L7M1A4 F6XU00 A0A310SK47 A0A088A1E3 V5HDR6 A0A2I2UL78 A0A1S4E770 A0A131YFK5 A0A0L7QVY0 A0A182Q9E5 A0A1D2NFJ1 A0A1Y1KCC5 E2C2S4 B4MVX2 A0A2Y9GHU4 A0A2U3WKS5 A0A2U3YZD9 A0A0M9A4P4 A0A2A3EIH6 R0LK38 A0A2P6LB90 M3XZJ6 U3IYE3 A0A224XFU0 A0A0P5MQY5 Q16HW6 A0A0P6DQT1 A0A182VUJ8 A0A1S4G0A0 A0A0P6HB29 A0A0P6FGV9 A0A0N8DJW2 G3MSF5 A0A023GDZ4 A0A0P5ZSE6 A0A182VAK9 Q7PVE7 A0A182T103 A0A1S4H1I0 A0A1Z5LAN5 G1LNH7 A0A226F3Q2 D2I2M6 W5J8Y5 B3N716 A0A0Q5WBM3 A0A182LY21 A0A182MZ21 F1LBC3 A0A232F6G9 T1EB03 A0A093LAM1 A0A1Z5L2V3 A0A0K8V4N5 A0A094LG59 A0A2Y9IRJ0 L5LW21 A0A212F733 A0A182P9G5 B4Q6B2 A0A091MPY0 A0A2M4BRP8 A0A0J9QYE7 B3MPI5 F1KS81 A0A0K8TT47 A0A2M4BRB8 Q9VLR9 Q9VLR9-2 K7IQV4 A0A034WAC4 S7MXE3 A0A2R2MIX8 A0A0P6J2Q9 A0A099ZM14 F4WK18 A0A3L8DLL8 A0A0B6ZV66 G1PHL5 A0A2Y9IVG2
Ontologies
GO
GO:0006506
GO:0016021
GO:0016787
GO:0051301
GO:0004527
GO:0005654
GO:0005794
GO:0008081
GO:0006888
GO:0005793
GO:0005783
GO:0034235
GO:0033116
GO:0070971
GO:0030145
GO:0005801
GO:0006044
GO:0005975
GO:0004342
GO:0046872
GO:0005737
GO:0006629
GO:0016627
GO:0004889
GO:0005216
GO:0045211
GO:0006281
GO:0048384
Topology
Subcellular location
Golgi apparatus
Also localizes to endoplasmic reticulum exit site. With evidence from 1 publications.
cis-Golgi network membrane Also localizes to endoplasmic reticulum exit site. With evidence from 1 publications.
Endoplasmic reticulum-Golgi intermediate compartment membrane Also localizes to endoplasmic reticulum exit site. With evidence from 1 publications.
Membrane
cis-Golgi network membrane Also localizes to endoplasmic reticulum exit site. With evidence from 1 publications.
Endoplasmic reticulum-Golgi intermediate compartment membrane Also localizes to endoplasmic reticulum exit site. With evidence from 1 publications.
Membrane
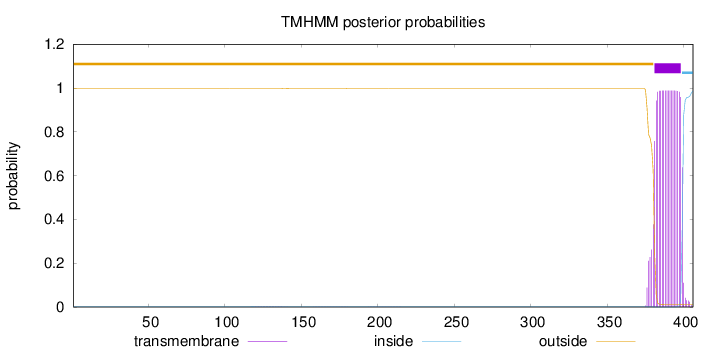
Length:
406
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.56779
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00305
outside
1 - 380
TMhelix
381 - 398
inside
399 - 406
Population Genetic Test Statistics
Pi
21.862011
Theta
23.621268
Tajima's D
-2.0807
CLR
62.150713
CSRT
0.0100494975251237
Interpretation
Uncertain
