Gene
KWMTBOMO02776
Pre Gene Modal
BGIBMGA003703
Annotation
PREDICTED:_cilia-_and_flagella-associated_protein_57_isoform_X2_[Amyelois_transitella]
Full name
Cilia- and flagella-associated protein 57
Alternative Name
WD repeat-containing protein 65
Location in the cell
Cytoplasmic Reliability : 2.095 Nuclear Reliability : 2.009
Sequence
CDS
ATGGCCCTGAATTCGCCGCCGAATCTTTCGGCTCGTATATTTTATGGTCTCCGCACAGATATTCAATATAATGCGCACTATTTATCGGAATCTGAAATCATCTACCCTGCTGGGGGAGTAATCGTTATTCACGACCATCTACAAAAGAAACAAAAATTTATTCGATTACAAGACAAACATAAACCAATTAAATCTCTGGTACTGGCACCTAACAGGCGTTGGCTTGCGCTAAATGAAATTGCCGAAGAGGGTCAAAAACCTATTATTACCATCTATGACCTAACTACGTACAAGAGACGTAAGATCCTAACGGTTCCATTCGAGAATTCGACCGCAAGAGAATTCGCTTGCATCCAGTTCACGTTTGATTCTAAATACCTCGTGGCAATAACGGGAGAACCAGATTGGTGCTTGTATTATTACAATTGGGACAAAGGAAAAGTTGAAAGTCATGCTAAAGCTCAAAATCCAAGTGGGCAAGGGACCGTTGAAAGCGTGCAATGTAATCCATCAGACGCTACTTTGGTGGTGATCACGGGACCGTACACATTTCGTATCATGAATGTTTCTGAAACTGTCTGGCGCCAGTGGGGTTGGTGTAAAGCCGAGAATATCGCCATAACAAGTTGTATGTGGCTCACACCAGATCGAATCTTGTTCGGCACTGAGAATGGCGTCATCATGATGGTCGAAAATGGTGAACTACGCCAAAATTGCATATTTCGAGCAGCTGAAGTAACAGAATTTTCGTTGAAGAAAGTAGAGGCCGAGGCAGAGGAAGGCGACAAGACAAGCACTACGAGTAAACAAGATAGCACGAGTGATTCAACAGTCTCTTTAGATGAAAGCCAGCCAGTTACTTGTTTTGTAAACTTTGCGAAAGGCTTTGCATATGCTTGTGGCCCCGGCTACGTACACATGTTTGAGAAGGAAACTCCTCATCACTGGAGGAAAAGAAACTTATTTAGAATTTCGAGAAAGTCTTATAAACATACCCGAGAACACCCTGTATGGTCAAATTTGGATGCAATACAACACATCACTATCGATCCCAACCAAGAAACTCTGCTCATTACAACGTTGCGAAAACAGCTGTATCATGTTAAATTATTTGGACAACATATGCTTCAAAATCCGGAAATACCTTTCACAGAATTAGGTCCGTCGATGCATTACGGCCCTATAAATTCACTGTCGATGTGTGCTTGGAAGCCCATATTTATGACATCTGGGGAGCAAGATAAAAGCATCAGGATATGGAATTATATGACTGACGACGTGGAAATGATCAAATTGTACCAGGAAGAAATCCATTGCGTGTCTCTGCACCCGGCTGGTCTATTCGCAATCGTAGGATTTTCAGACAAGCTTCGTTTCATGGTTGTGCTGATTGATGATTTTGAAGTGATGCGGGAATTCCCCATACGTAATTGCAAACAAGCAAAATTTAGCACGAACGGTCACCTTTTTGCAGCAGTAAATGGCCAAGTGATCCAAGTTTTTTCCTCAGTTTCATTTCAAAATGTTTATAATTTAAAAGGACACAATGGGAGAATAACGTGTTTGGCTTGGTCAGCTAACGACTTGACTTTAGTATCTTGCGGCACGGAAGGCGCCGTTTACGAATGGAGTATGTCGTCTGGGCAAAGAGTCGGAGAAGTTATACTAAAAACGAATCAGTTTAAAGCATGTGCAGTGAATAGTATCGGAAAAACTACGTACGGTCTCGGAACTGATGGAGAAATAAAGGAAATCGGATCAAACACTATCAGACGGAATTTGGGTTTGATTGGATGTGCTCTTGATACGATCGTGTTATCGCGTTCCGATATGATGCTTTTTGTAACCGGTGGTGATGGAGGTGTAACGTCAGTGCAGTTGCCGCTCCTCGACAAGGCTGTTTTTAATGAGTTTCACATGCATAATAAAAAAGTAACTTGCATAGCTCTAGCTTACGACGATCAAACATTAGTTTCTGTAGCTGAAGACGCTTCCATTTGCTTATGGAGACTCACGAACGCTGATGGAAGAGCTATTGCCCTTGAGAAAGATTTCGCATACTCCAAGGAGATTCTTATTAGTAAAAAAGATCTTCAAGAAAAAATTAATAATATCAACTTGTTGAGTACAAGGATGAGCGAATTGGAAACGGAGCACACATATCAGCTTCGTCAAGCTGAAGCAACACAAGCCGAAAAACTGAAAGAAGTCCACGAAGGTTATTGCGCTGCAATAGAGGAATTAAAAGAAAAGAACGAGCTAATGGAAAGTGAACATACGCACGAAATAGGTATGATTCAACAAGATATTGCAAAATTGCGGTCTGGTCACGAGCGAACATTGCAGGCGCTCGAAGCGGATTTCAATGTCAGACTTATTAACGAGTACGACAGATACCAGAGTCTGGAAGACAAAACCGCCCGAATGCGAAAAGATTACGAAGAACGGCTCGAGGAATTAGCTGAAAGTAAGCTTCAAGCTCTTCGAGAAATTAATAATATGTTTGAAGCTAAACTCGAAGAAAAGGATTTACTCTTACAAGAGTTACAAGAACAAACAGACTTAGAAAGACGTGAACATGAAACAATAAAGGCGTCTATTGAAGAGGATGCTGATCGAGAAATTATTGAAATAAGAACGGCTTATGAAGTTCAATTGAAAGAGGAAAAGGACGCTAACGTTAGGTTAAAAGGTGAAACTGGACTTATGAAAAAGAAATTAATTAGTGCTAATAAAGAAATAGACGAGTTTAAGCATCAGGTTTCACAACTCAAAGCTGAACACAAACAGTTTCAAAAGGTAATATCAACATTGGAACGAGATGTGGCAGACTTGAAAAAAGAGATTTCAGAAAGAGATGGAACAATACAAGATAAAGAGAAACGAATATATGAATTGAAAAGGAAAAAGCAGGAATTAGAGAAGTACAAGTTTGTGCTTAATTTTAAAATAACCGAACTTAAAAATCAAATTGAGCCAAAAGAACGTATAATAAGTGAGCTTAAAGTTCAGATCGATGACATGGAAAACGAAGAATTAAAGCTGTTGAACGTTAAACATGAACTTGAGCTAAAAATCAGCCAGTTAAACGAAAAATTAGCATCGGCCAAAAAGGATTACTTAAATGAAGCCGACAGGAATGCAATGCTGAAGAACACATTAAAAAAAATAAAGATTGATTTGCATAATATGACGGCAAATTTTCAAGATCCAATGAAATTAAAAAGTAATGTGAAGACACTGTTTCACAAATACGTTGAAGACATCGATTTCGTACGCAGCCGTGTGGCAGAGGATGAAGCTATACGTGAATTCAACAGGCAGAGAGATCACCTTGAAAAACAGGTCACAGCTCTCAAACAACAATTAGCAAAGTCACTTGGTGGATCCAAGAGCGATATCGGAAAAATTATGGACGAAAACTGCACTCTACTGGCTGAGATCAATAGTCTAAGAACAGAGTTAAAATCGACTCGTTCTAGATGTTTTCAAATGGAGTCAATGCTTGGCTTGTCAGCGCGGTACATACCGCCAGCCACGGCTCGAGCTAAACTTAAGCATGTCACCGAAGATCGCGATAAACTTGATGAAGGCTTTAAGCAGAAAATTGAGGAAAGAGAAGAGATAATCATTGCTTTGAAAGAAGAGAATGAACGTCTATTAGGTAAAATACGATGTGTAGAAAATTCGGGGACAGCTACAGAAGATCCAGACGGGGCTAAAAATGAAATGTATTAA
Protein
MALNSPPNLSARIFYGLRTDIQYNAHYLSESEIIYPAGGVIVIHDHLQKKQKFIRLQDKHKPIKSLVLAPNRRWLALNEIAEEGQKPIITIYDLTTYKRRKILTVPFENSTAREFACIQFTFDSKYLVAITGEPDWCLYYYNWDKGKVESHAKAQNPSGQGTVESVQCNPSDATLVVITGPYTFRIMNVSETVWRQWGWCKAENIAITSCMWLTPDRILFGTENGVIMMVENGELRQNCIFRAAEVTEFSLKKVEAEAEEGDKTSTTSKQDSTSDSTVSLDESQPVTCFVNFAKGFAYACGPGYVHMFEKETPHHWRKRNLFRISRKSYKHTREHPVWSNLDAIQHITIDPNQETLLITTLRKQLYHVKLFGQHMLQNPEIPFTELGPSMHYGPINSLSMCAWKPIFMTSGEQDKSIRIWNYMTDDVEMIKLYQEEIHCVSLHPAGLFAIVGFSDKLRFMVVLIDDFEVMREFPIRNCKQAKFSTNGHLFAAVNGQVIQVFSSVSFQNVYNLKGHNGRITCLAWSANDLTLVSCGTEGAVYEWSMSSGQRVGEVILKTNQFKACAVNSIGKTTYGLGTDGEIKEIGSNTIRRNLGLIGCALDTIVLSRSDMMLFVTGGDGGVTSVQLPLLDKAVFNEFHMHNKKVTCIALAYDDQTLVSVAEDASICLWRLTNADGRAIALEKDFAYSKEILISKKDLQEKINNINLLSTRMSELETEHTYQLRQAEATQAEKLKEVHEGYCAAIEELKEKNELMESEHTHEIGMIQQDIAKLRSGHERTLQALEADFNVRLINEYDRYQSLEDKTARMRKDYEERLEELAESKLQALREINNMFEAKLEEKDLLLQELQEQTDLERREHETIKASIEEDADREIIEIRTAYEVQLKEEKDANVRLKGETGLMKKKLISANKEIDEFKHQVSQLKAEHKQFQKVISTLERDVADLKKEISERDGTIQDKEKRIYELKRKKQELEKYKFVLNFKITELKNQIEPKERIISELKVQIDDMENEELKLLNVKHELELKISQLNEKLASAKKDYLNEADRNAMLKNTLKKIKIDLHNMTANFQDPMKLKSNVKTLFHKYVEDIDFVRSRVAEDEAIREFNRQRDHLEKQVTALKQQLAKSLGGSKSDIGKIMDENCTLLAEINSLRTELKSTRSRCFQMESMLGLSARYIPPATARAKLKHVTEDRDKLDEGFKQKIEEREEIIIALKEENERLLGKIRCVENSGTATEDPDGAKNEMY
Summary
Keywords
Alternative splicing
Coiled coil
Complete proteome
Polymorphism
Reference proteome
Repeat
WD repeat
Feature
chain Cilia- and flagella-associated protein 57
splice variant In isoform 2.
sequence variant In dbSNP:rs6663799.
splice variant In isoform 2.
sequence variant In dbSNP:rs6663799.
Uniprot
H9J2G6
A0A212ETB2
A0A194QQ18
A0A2H1WLQ3
A0A194PUT3
A0A2A4JXX9
+ More
A0A2J7R3M8 D6WVH5 E1ZVD5 E9IFJ3 A0A026W6V1 A0A151X9E4 A0A0J7N9G5 A0A195DRZ0 A0A195BG73 A0A158NC20 A0A195FD87 A0A1B6LPR0 A0A151IMB9 K7IQG5 E2BC92 A0A232F5D2 A0A0L7R1U0 A0A088A5V7 A0A2A3EIY3 F4X878 A0A0C9RK59 U4UED5 A0A1W5BM36 A0A2C9K1G6 A0A2T7NNI4 A0A210Q048 A0A1L8GM02 A0A0B7A6G0 A0A1S3IR34 W5MH95 W5MH82 N6TXR1 A0A1W4ZYJ0 A0A369SH58 W5QH30 K1QFI0 M3YQQ8 A0A2Y9J4E3 A0A3P8XHD0 A0A384CST8 H0Y248 A0A2Y9GJW5 A0A2U3VYR0 E2QWR4 H2ZKC6 A0A3P8ZJA3 A0A2Y9NPC6 A0A1U7TEK8 M3XAG8 A0A267F950 A0A1I8IX27 A0A267F1I4 A0A383YPH6 A0A1I8GKQ9 G1KNF2 F6TVM2 A0A1U7R9B0 A0A1S3WF92 A0A341BE64 A0A2K6FMT4 G3HFH7 G3TLA4 S7MT86 A0A1S3QXT4 A0A1S3QXT0 F6S6M6 A0A1S3F7J4 A0A2J8SME3 A0A2K6K3S6 A0A2K6NKI3 F7H742 A0A337SJ05 A0A2K5J6R0 A0A151MKT8 A0A369SH67 A0A0D9S7J7 A0A074ZBK4 A0A2Y9DCA2 H0VA87 A0A2Y9SS87 A0A2R9B388 H2PYU5 A0A1A6GXH0 Q96MR6 G1SN80 G3S7U2 A0A340Y7H8 V8PHE8 A0A2K5EK89 A0A3B4CKV7 A0A2K6K3V2 A0A2K6FMU8 F7GKM5
A0A2J7R3M8 D6WVH5 E1ZVD5 E9IFJ3 A0A026W6V1 A0A151X9E4 A0A0J7N9G5 A0A195DRZ0 A0A195BG73 A0A158NC20 A0A195FD87 A0A1B6LPR0 A0A151IMB9 K7IQG5 E2BC92 A0A232F5D2 A0A0L7R1U0 A0A088A5V7 A0A2A3EIY3 F4X878 A0A0C9RK59 U4UED5 A0A1W5BM36 A0A2C9K1G6 A0A2T7NNI4 A0A210Q048 A0A1L8GM02 A0A0B7A6G0 A0A1S3IR34 W5MH95 W5MH82 N6TXR1 A0A1W4ZYJ0 A0A369SH58 W5QH30 K1QFI0 M3YQQ8 A0A2Y9J4E3 A0A3P8XHD0 A0A384CST8 H0Y248 A0A2Y9GJW5 A0A2U3VYR0 E2QWR4 H2ZKC6 A0A3P8ZJA3 A0A2Y9NPC6 A0A1U7TEK8 M3XAG8 A0A267F950 A0A1I8IX27 A0A267F1I4 A0A383YPH6 A0A1I8GKQ9 G1KNF2 F6TVM2 A0A1U7R9B0 A0A1S3WF92 A0A341BE64 A0A2K6FMT4 G3HFH7 G3TLA4 S7MT86 A0A1S3QXT4 A0A1S3QXT0 F6S6M6 A0A1S3F7J4 A0A2J8SME3 A0A2K6K3S6 A0A2K6NKI3 F7H742 A0A337SJ05 A0A2K5J6R0 A0A151MKT8 A0A369SH67 A0A0D9S7J7 A0A074ZBK4 A0A2Y9DCA2 H0VA87 A0A2Y9SS87 A0A2R9B388 H2PYU5 A0A1A6GXH0 Q96MR6 G1SN80 G3S7U2 A0A340Y7H8 V8PHE8 A0A2K5EK89 A0A3B4CKV7 A0A2K6K3V2 A0A2K6FMU8 F7GKM5
Pubmed
19121390
22118469
26354079
18362917
19820115
20798317
+ More
21282665 24508170 30249741 21347285 20075255 28648823 21719571 23537049 15562597 28812685 27762356 30042472 20809919 22992520 25069045 16341006 17975172 26392545 19892987 21804562 29704459 17495919 25362486 17431167 22293439 21993624 22722832 16136131 14702039 16710414 15489334 21574244 22398555 24297900
21282665 24508170 30249741 21347285 20075255 28648823 21719571 23537049 15562597 28812685 27762356 30042472 20809919 22992520 25069045 16341006 17975172 26392545 19892987 21804562 29704459 17495919 25362486 17431167 22293439 21993624 22722832 16136131 14702039 16710414 15489334 21574244 22398555 24297900
EMBL
BABH01007930
AGBW02012631
OWR44691.1
KQ461181
KPJ07608.1
ODYU01009501
+ More
SOQ53989.1 KQ459597 KPI94895.1 NWSH01000406 PCG76626.1 NEVH01007818 PNF35437.1 KQ971357 EFA08298.1 GL434492 EFN74845.1 GL762841 EFZ20654.1 KK107372 QOIP01000004 EZA51703.1 RLU23506.1 KQ982373 KYQ56991.1 LBMM01007908 KMQ89315.1 KQ980581 KYN15284.1 KQ976500 KYM83204.1 ADTU01011290 KQ981673 KYN38346.1 GEBQ01014226 JAT25751.1 KQ977059 KYN06041.1 GL447257 EFN86628.1 NNAY01000969 OXU25673.1 KQ414667 KOC64830.1 KZ288229 PBC31680.1 GL888932 EGI57209.1 GBYB01013667 GBYB01013672 JAG83434.1 JAG83439.1 KB632283 ERL91397.1 PZQS01000010 PVD22724.1 NEDP02005318 OWF42120.1 CM004472 OCT84882.1 HACG01028745 HACG01028746 CEK75610.1 CEK75611.1 AHAT01015953 APGK01057208 APGK01057209 APGK01057210 APGK01057211 APGK01057212 APGK01057213 APGK01057214 APGK01057215 APGK01057216 KB741278 ENN71062.1 NOWV01000010 RDD46258.1 AMGL01001101 AMGL01001102 AMGL01001103 JH816435 EKC35627.1 AEYP01007510 AEYP01007511 AEYP01007512 AAQR03170722 AAQR03170723 AAQR03170724 AAQR03170725 AAQR03170726 AAQR03170727 AAEX03009815 AAEX03009816 AANG04003279 NIVC01001296 PAA69734.1 NIVC01001472 PAA67586.1 JH000327 RAZU01000087 EGW08128.1 RLQ74201.1 KE162280 EPQ07674.1 NDHI03003559 PNJ21946.1 JSUE03001239 AKHW03005990 KYO25063.1 RDD46257.1 AQIB01144757 AQIB01144758 AQIB01144759 AQIB01144760 AQIB01144761 AQIB01144762 KL597039 KER20605.1 AAKN02053650 AJFE02040935 AJFE02040936 AJFE02040937 AJFE02040938 AJFE02040939 AACZ04033001 NBAG03000275 PNI52630.1 LZPO01066267 OBS70856.1 AK056562 AC093420 AL139138 BC117280 BC117306 AAGW02003011 AAGW02003012 AAGW02003013 AAGW02003014 CABD030002802 CABD030002803 CABD030002804 AZIM01000100 ETE73386.1
SOQ53989.1 KQ459597 KPI94895.1 NWSH01000406 PCG76626.1 NEVH01007818 PNF35437.1 KQ971357 EFA08298.1 GL434492 EFN74845.1 GL762841 EFZ20654.1 KK107372 QOIP01000004 EZA51703.1 RLU23506.1 KQ982373 KYQ56991.1 LBMM01007908 KMQ89315.1 KQ980581 KYN15284.1 KQ976500 KYM83204.1 ADTU01011290 KQ981673 KYN38346.1 GEBQ01014226 JAT25751.1 KQ977059 KYN06041.1 GL447257 EFN86628.1 NNAY01000969 OXU25673.1 KQ414667 KOC64830.1 KZ288229 PBC31680.1 GL888932 EGI57209.1 GBYB01013667 GBYB01013672 JAG83434.1 JAG83439.1 KB632283 ERL91397.1 PZQS01000010 PVD22724.1 NEDP02005318 OWF42120.1 CM004472 OCT84882.1 HACG01028745 HACG01028746 CEK75610.1 CEK75611.1 AHAT01015953 APGK01057208 APGK01057209 APGK01057210 APGK01057211 APGK01057212 APGK01057213 APGK01057214 APGK01057215 APGK01057216 KB741278 ENN71062.1 NOWV01000010 RDD46258.1 AMGL01001101 AMGL01001102 AMGL01001103 JH816435 EKC35627.1 AEYP01007510 AEYP01007511 AEYP01007512 AAQR03170722 AAQR03170723 AAQR03170724 AAQR03170725 AAQR03170726 AAQR03170727 AAEX03009815 AAEX03009816 AANG04003279 NIVC01001296 PAA69734.1 NIVC01001472 PAA67586.1 JH000327 RAZU01000087 EGW08128.1 RLQ74201.1 KE162280 EPQ07674.1 NDHI03003559 PNJ21946.1 JSUE03001239 AKHW03005990 KYO25063.1 RDD46257.1 AQIB01144757 AQIB01144758 AQIB01144759 AQIB01144760 AQIB01144761 AQIB01144762 KL597039 KER20605.1 AAKN02053650 AJFE02040935 AJFE02040936 AJFE02040937 AJFE02040938 AJFE02040939 AACZ04033001 NBAG03000275 PNI52630.1 LZPO01066267 OBS70856.1 AK056562 AC093420 AL139138 BC117280 BC117306 AAGW02003011 AAGW02003012 AAGW02003013 AAGW02003014 CABD030002802 CABD030002803 CABD030002804 AZIM01000100 ETE73386.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000235965
+ More
UP000007266 UP000000311 UP000053097 UP000279307 UP000075809 UP000036403 UP000078492 UP000078540 UP000005205 UP000078541 UP000078542 UP000002358 UP000008237 UP000215335 UP000053825 UP000005203 UP000242457 UP000007755 UP000030742 UP000076420 UP000245119 UP000242188 UP000186698 UP000085678 UP000018468 UP000019118 UP000192224 UP000253843 UP000002356 UP000005408 UP000000715 UP000248482 UP000265140 UP000261680 UP000005225 UP000248481 UP000245340 UP000002254 UP000007875 UP000248483 UP000189704 UP000011712 UP000215902 UP000095280 UP000261681 UP000001646 UP000002281 UP000189706 UP000079721 UP000252040 UP000233160 UP000001075 UP000273346 UP000007646 UP000087266 UP000002280 UP000081671 UP000233180 UP000233200 UP000006718 UP000233080 UP000050525 UP000029965 UP000248480 UP000005447 UP000248484 UP000240080 UP000002277 UP000092124 UP000005640 UP000001811 UP000001519 UP000265300 UP000233020 UP000261440 UP000008225
UP000007266 UP000000311 UP000053097 UP000279307 UP000075809 UP000036403 UP000078492 UP000078540 UP000005205 UP000078541 UP000078542 UP000002358 UP000008237 UP000215335 UP000053825 UP000005203 UP000242457 UP000007755 UP000030742 UP000076420 UP000245119 UP000242188 UP000186698 UP000085678 UP000018468 UP000019118 UP000192224 UP000253843 UP000002356 UP000005408 UP000000715 UP000248482 UP000265140 UP000261680 UP000005225 UP000248481 UP000245340 UP000002254 UP000007875 UP000248483 UP000189704 UP000011712 UP000215902 UP000095280 UP000261681 UP000001646 UP000002281 UP000189706 UP000079721 UP000252040 UP000233160 UP000001075 UP000273346 UP000007646 UP000087266 UP000002280 UP000081671 UP000233180 UP000233200 UP000006718 UP000233080 UP000050525 UP000029965 UP000248480 UP000005447 UP000248484 UP000240080 UP000002277 UP000092124 UP000005640 UP000001811 UP000001519 UP000265300 UP000233020 UP000261440 UP000008225
Pfam
PF00400 WD40
Interpro
Gene 3D
ProteinModelPortal
H9J2G6
A0A212ETB2
A0A194QQ18
A0A2H1WLQ3
A0A194PUT3
A0A2A4JXX9
+ More
A0A2J7R3M8 D6WVH5 E1ZVD5 E9IFJ3 A0A026W6V1 A0A151X9E4 A0A0J7N9G5 A0A195DRZ0 A0A195BG73 A0A158NC20 A0A195FD87 A0A1B6LPR0 A0A151IMB9 K7IQG5 E2BC92 A0A232F5D2 A0A0L7R1U0 A0A088A5V7 A0A2A3EIY3 F4X878 A0A0C9RK59 U4UED5 A0A1W5BM36 A0A2C9K1G6 A0A2T7NNI4 A0A210Q048 A0A1L8GM02 A0A0B7A6G0 A0A1S3IR34 W5MH95 W5MH82 N6TXR1 A0A1W4ZYJ0 A0A369SH58 W5QH30 K1QFI0 M3YQQ8 A0A2Y9J4E3 A0A3P8XHD0 A0A384CST8 H0Y248 A0A2Y9GJW5 A0A2U3VYR0 E2QWR4 H2ZKC6 A0A3P8ZJA3 A0A2Y9NPC6 A0A1U7TEK8 M3XAG8 A0A267F950 A0A1I8IX27 A0A267F1I4 A0A383YPH6 A0A1I8GKQ9 G1KNF2 F6TVM2 A0A1U7R9B0 A0A1S3WF92 A0A341BE64 A0A2K6FMT4 G3HFH7 G3TLA4 S7MT86 A0A1S3QXT4 A0A1S3QXT0 F6S6M6 A0A1S3F7J4 A0A2J8SME3 A0A2K6K3S6 A0A2K6NKI3 F7H742 A0A337SJ05 A0A2K5J6R0 A0A151MKT8 A0A369SH67 A0A0D9S7J7 A0A074ZBK4 A0A2Y9DCA2 H0VA87 A0A2Y9SS87 A0A2R9B388 H2PYU5 A0A1A6GXH0 Q96MR6 G1SN80 G3S7U2 A0A340Y7H8 V8PHE8 A0A2K5EK89 A0A3B4CKV7 A0A2K6K3V2 A0A2K6FMU8 F7GKM5
A0A2J7R3M8 D6WVH5 E1ZVD5 E9IFJ3 A0A026W6V1 A0A151X9E4 A0A0J7N9G5 A0A195DRZ0 A0A195BG73 A0A158NC20 A0A195FD87 A0A1B6LPR0 A0A151IMB9 K7IQG5 E2BC92 A0A232F5D2 A0A0L7R1U0 A0A088A5V7 A0A2A3EIY3 F4X878 A0A0C9RK59 U4UED5 A0A1W5BM36 A0A2C9K1G6 A0A2T7NNI4 A0A210Q048 A0A1L8GM02 A0A0B7A6G0 A0A1S3IR34 W5MH95 W5MH82 N6TXR1 A0A1W4ZYJ0 A0A369SH58 W5QH30 K1QFI0 M3YQQ8 A0A2Y9J4E3 A0A3P8XHD0 A0A384CST8 H0Y248 A0A2Y9GJW5 A0A2U3VYR0 E2QWR4 H2ZKC6 A0A3P8ZJA3 A0A2Y9NPC6 A0A1U7TEK8 M3XAG8 A0A267F950 A0A1I8IX27 A0A267F1I4 A0A383YPH6 A0A1I8GKQ9 G1KNF2 F6TVM2 A0A1U7R9B0 A0A1S3WF92 A0A341BE64 A0A2K6FMT4 G3HFH7 G3TLA4 S7MT86 A0A1S3QXT4 A0A1S3QXT0 F6S6M6 A0A1S3F7J4 A0A2J8SME3 A0A2K6K3S6 A0A2K6NKI3 F7H742 A0A337SJ05 A0A2K5J6R0 A0A151MKT8 A0A369SH67 A0A0D9S7J7 A0A074ZBK4 A0A2Y9DCA2 H0VA87 A0A2Y9SS87 A0A2R9B388 H2PYU5 A0A1A6GXH0 Q96MR6 G1SN80 G3S7U2 A0A340Y7H8 V8PHE8 A0A2K5EK89 A0A3B4CKV7 A0A2K6K3V2 A0A2K6FMU8 F7GKM5
PDB
1I84
E-value=1.61923e-06,
Score=129
Ontologies
GO
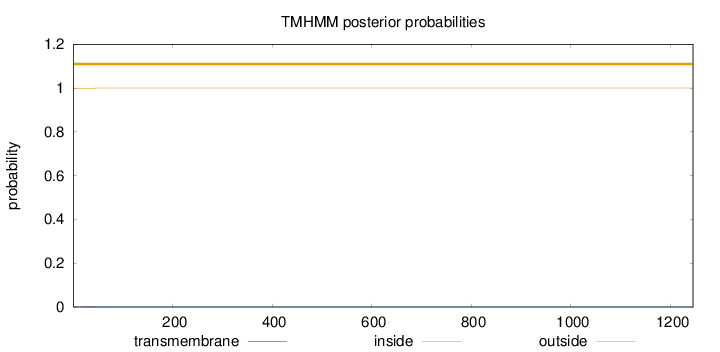
Topology
Length:
1245
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02348
Exp number, first 60 AAs:
0.02033
Total prob of N-in:
0.00097
outside
1 - 1245
Population Genetic Test Statistics
Pi
206.496218
Theta
174.069968
Tajima's D
-1.011158
CLR
24.01958
CSRT
0.134243287835608
Interpretation
Uncertain
